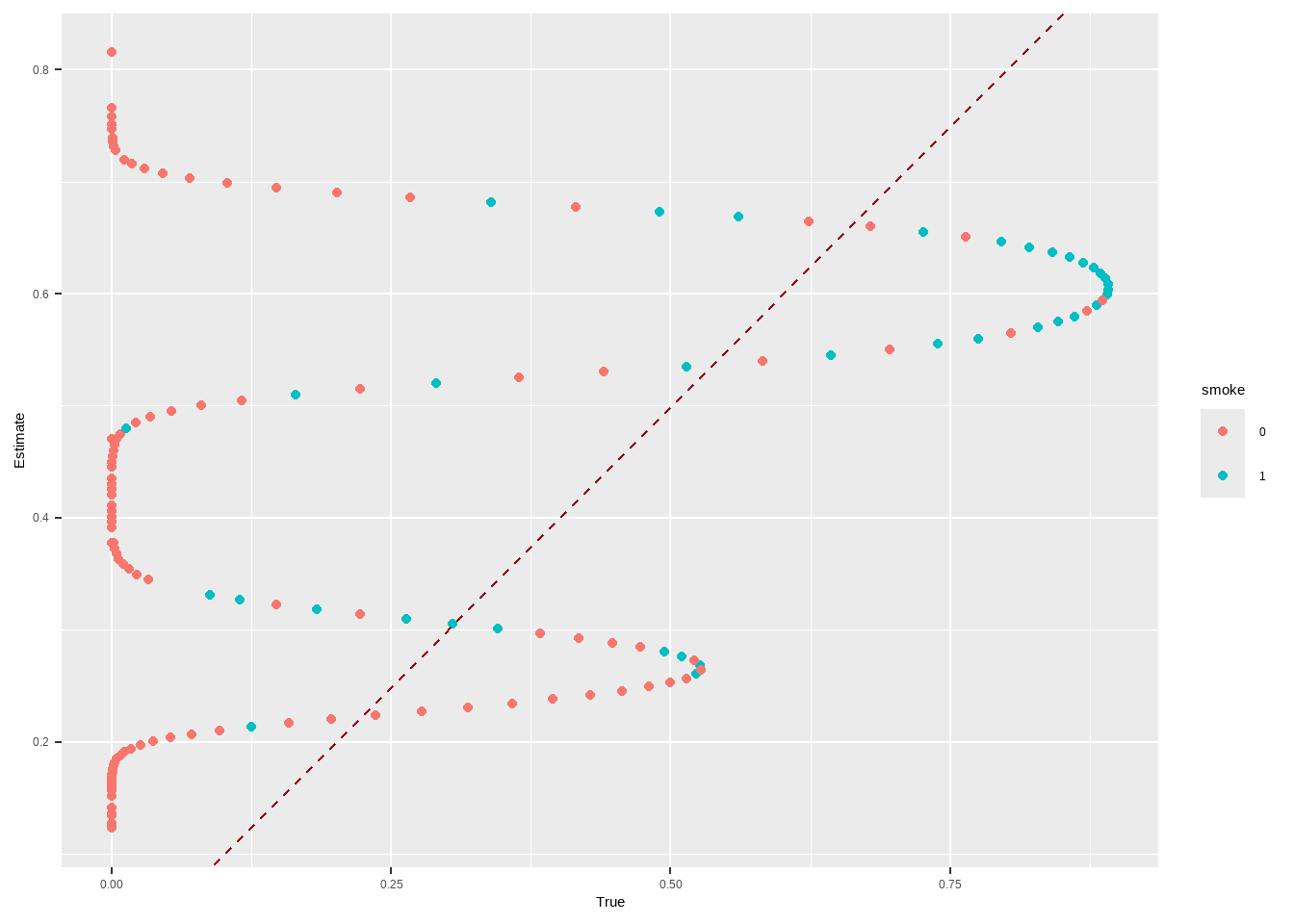
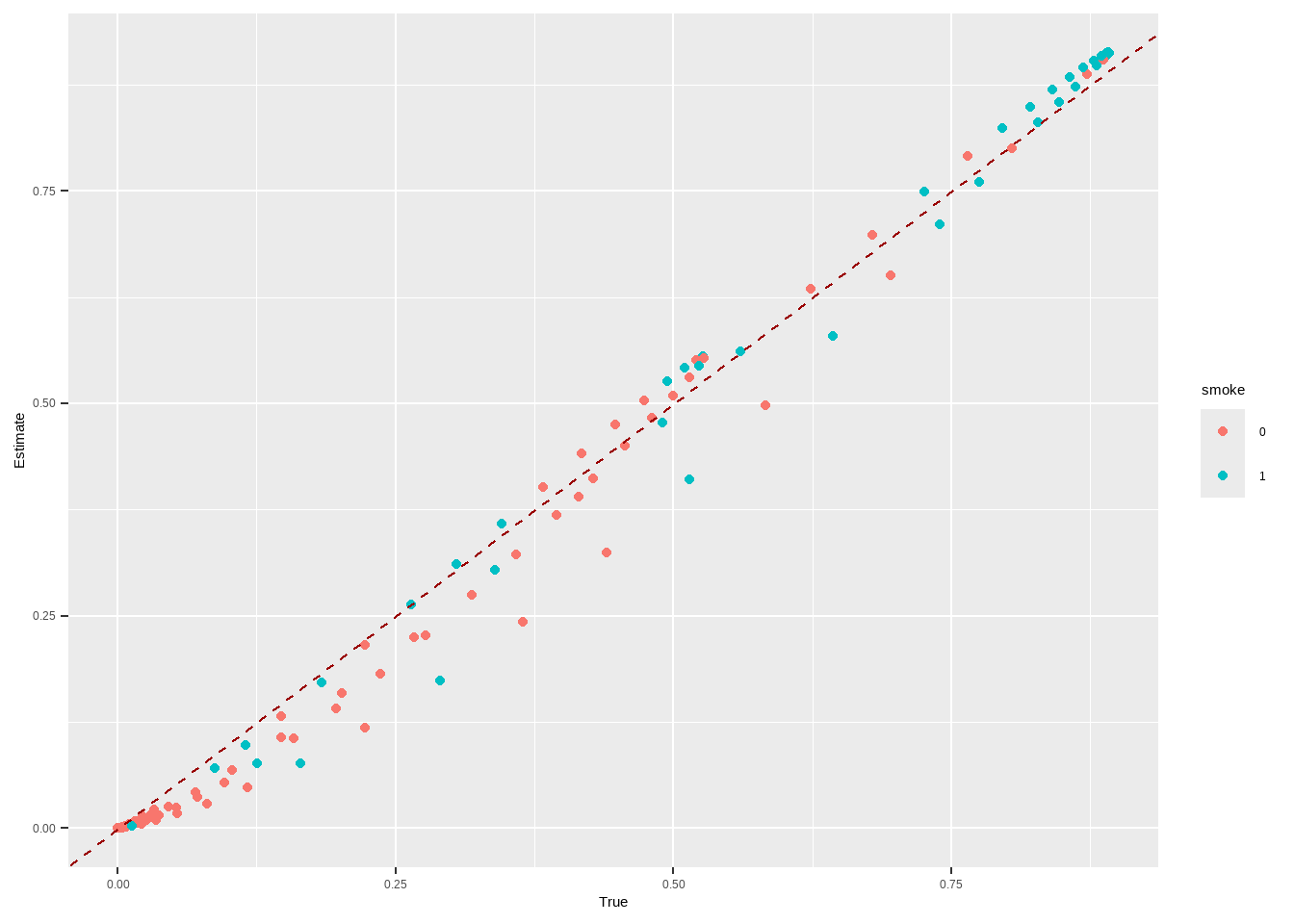
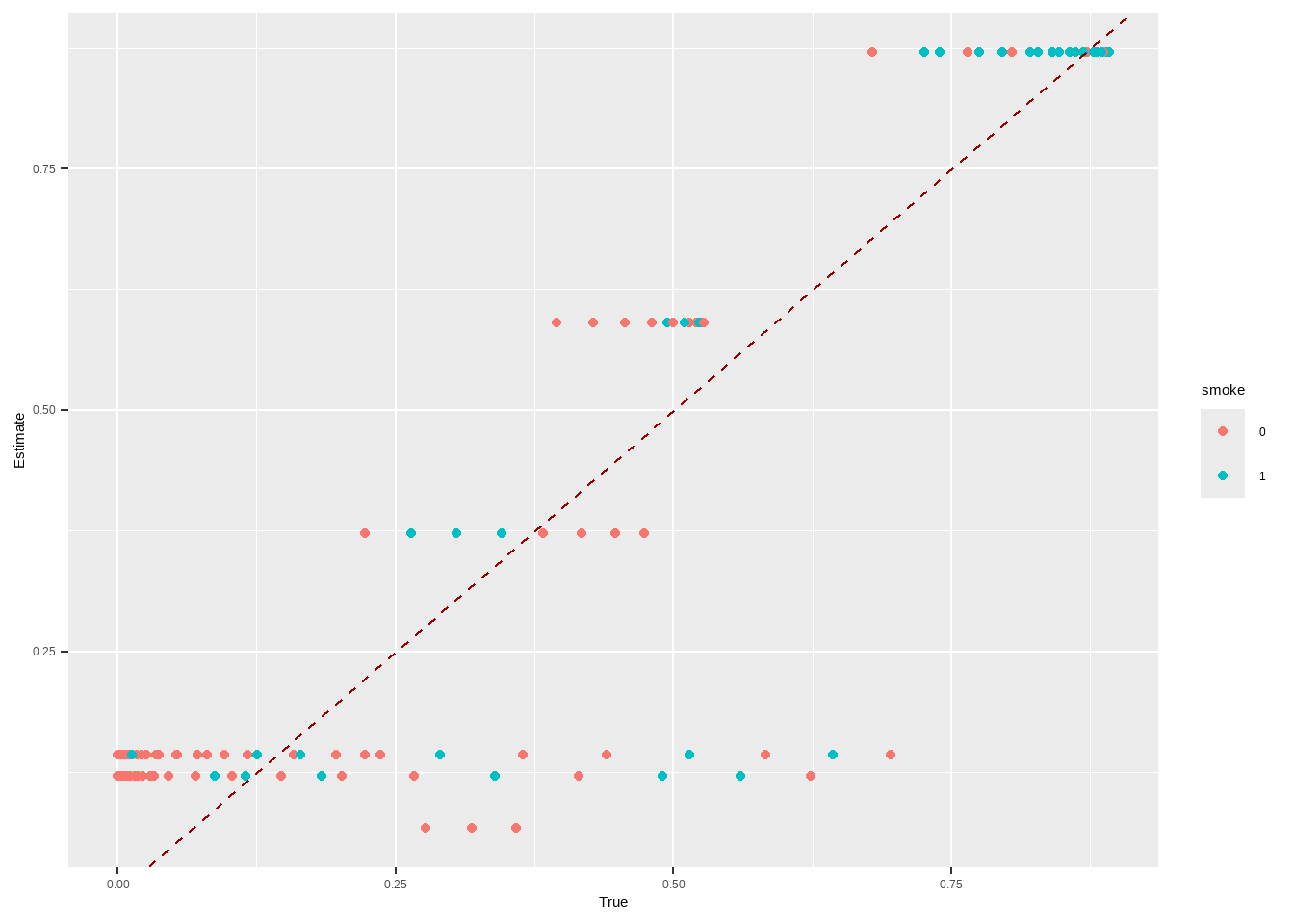
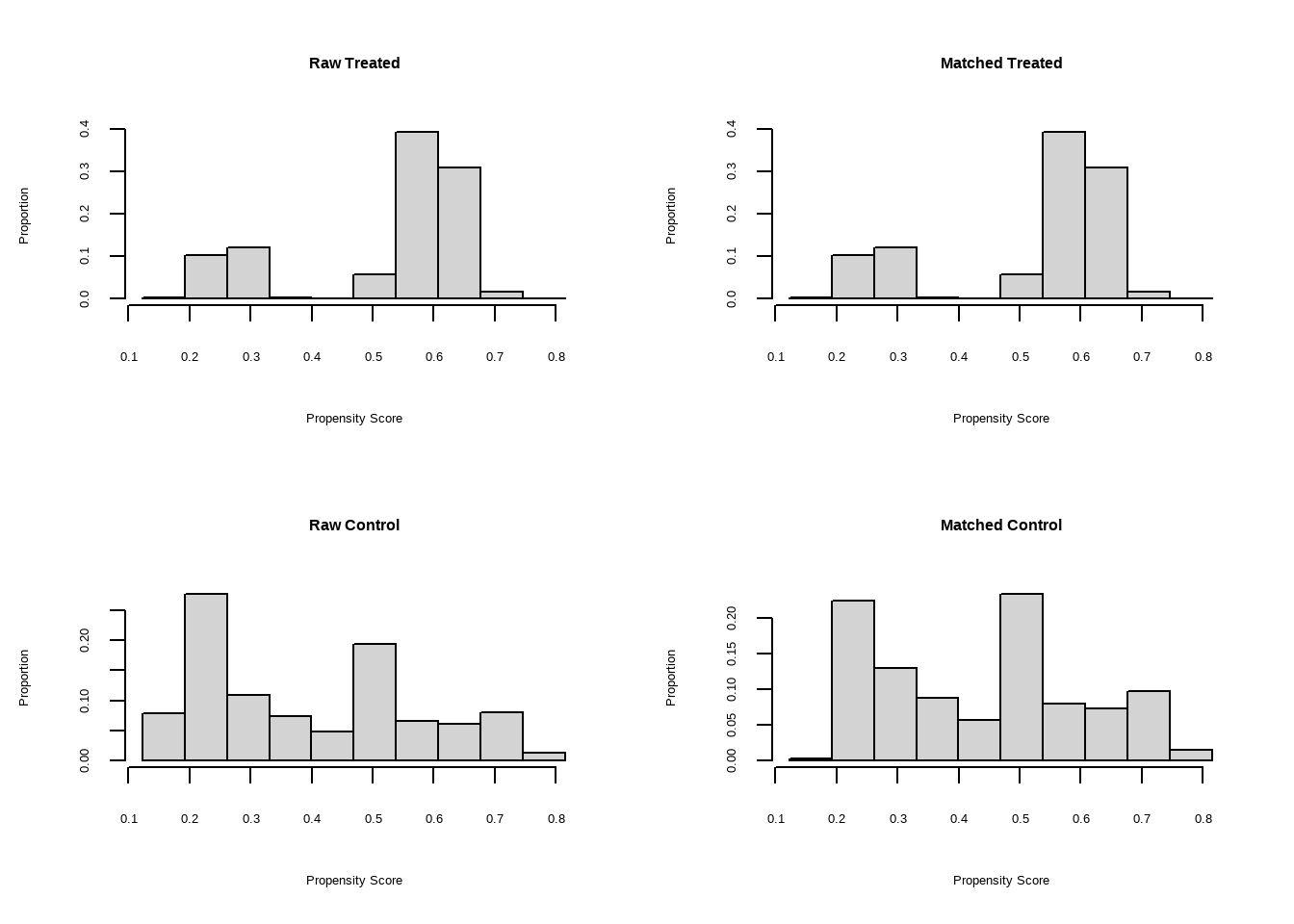
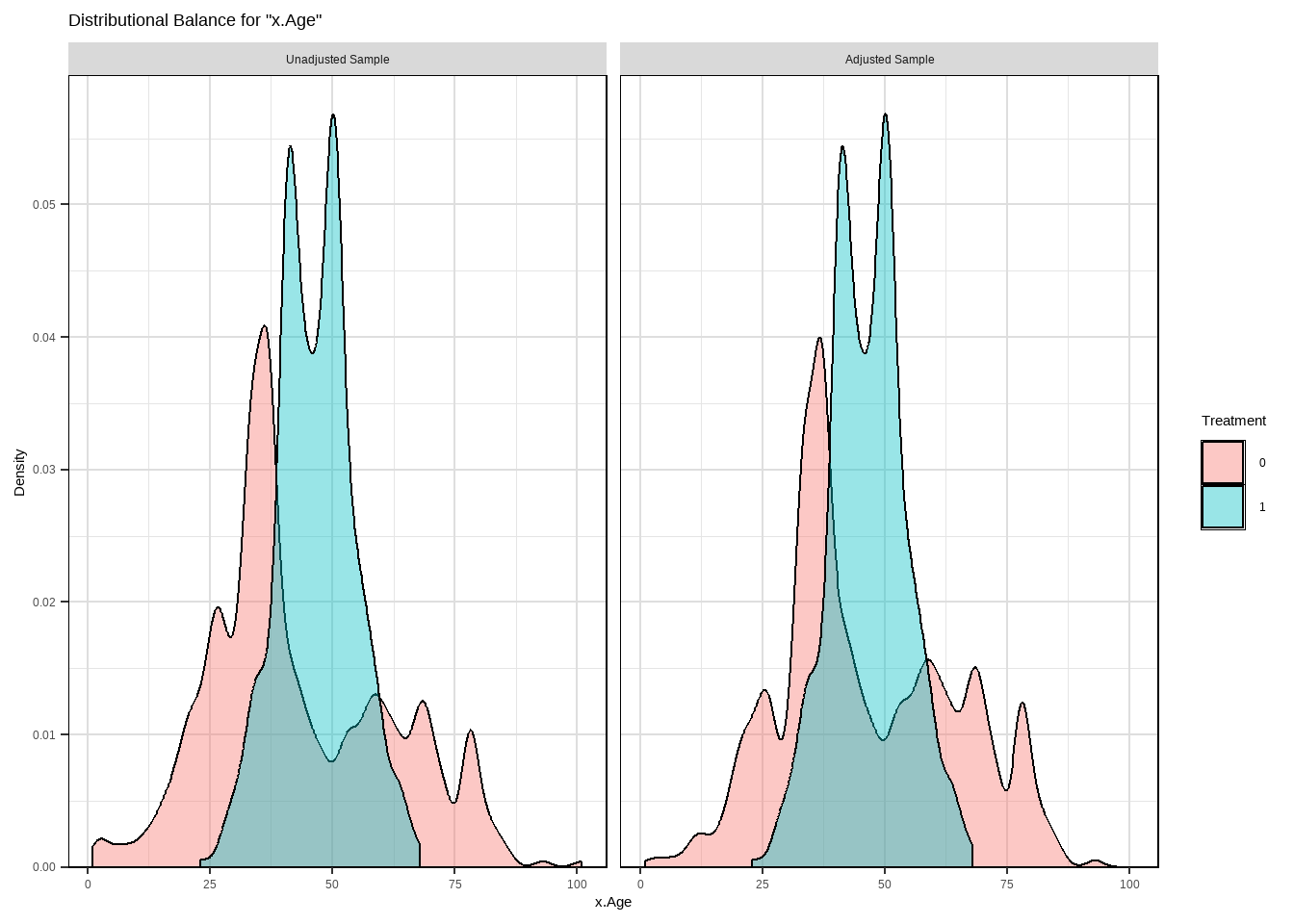
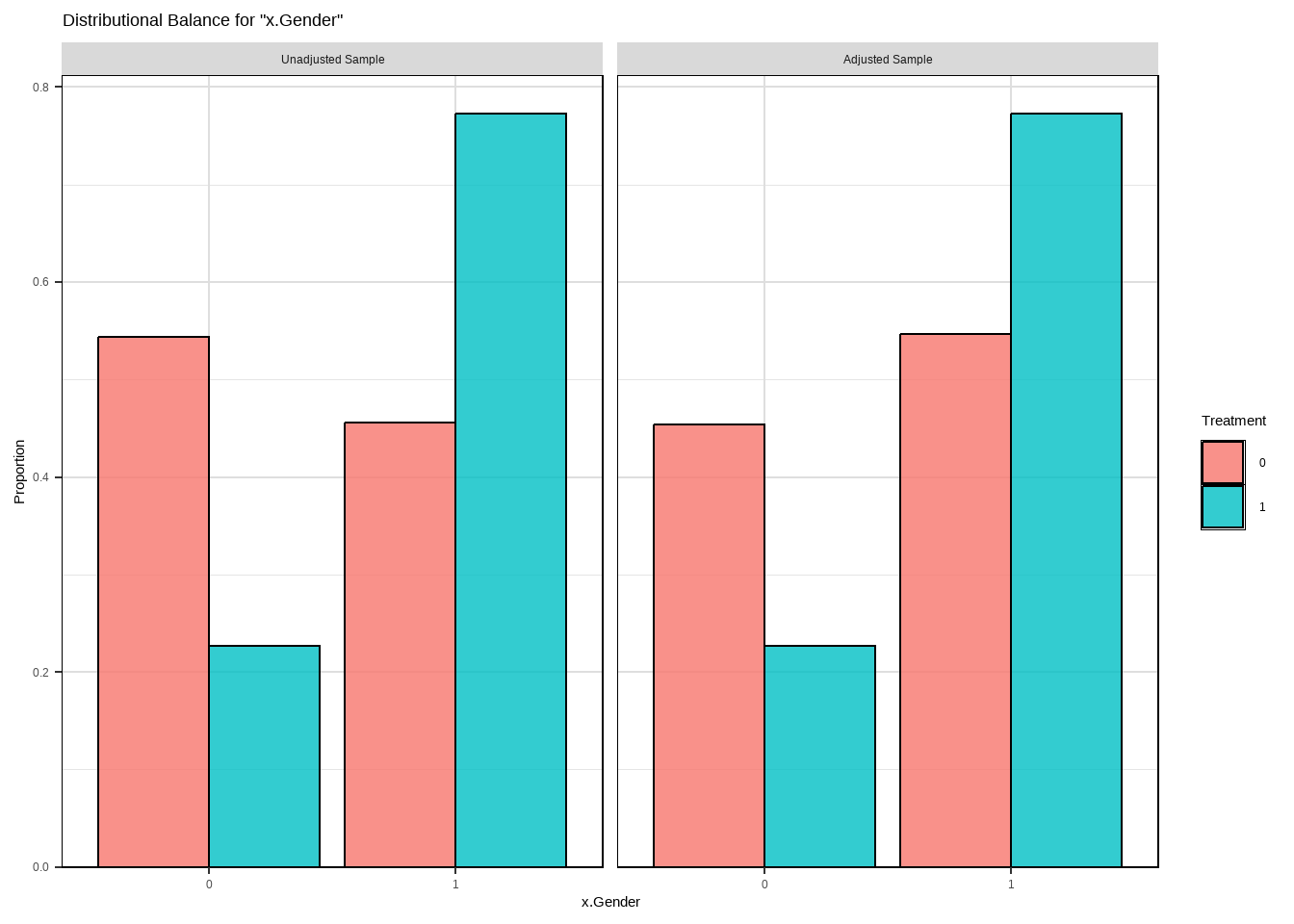
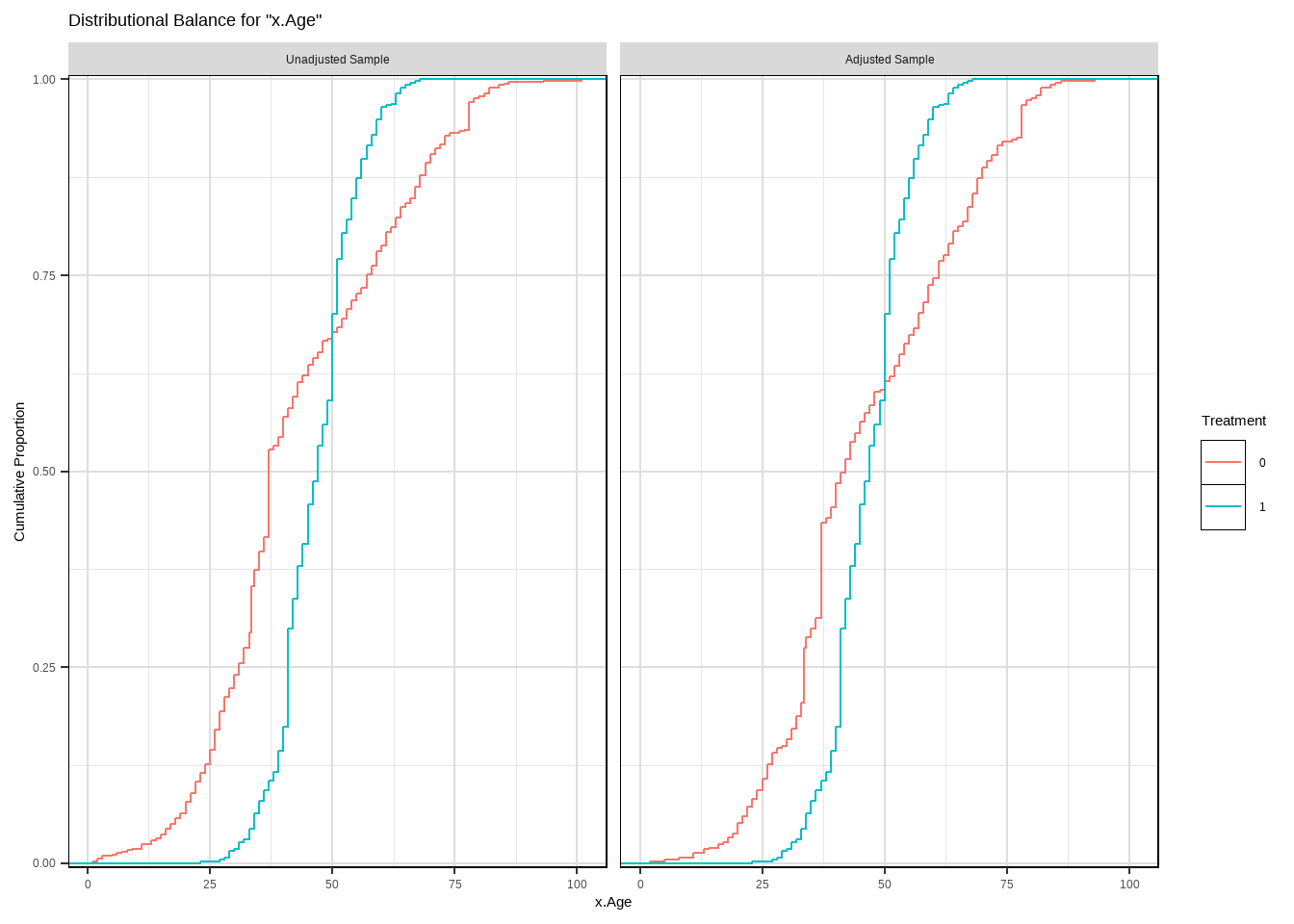
m.out2 $ model #> #> Call: glm(formula = Smoke ~ x.Age + I(x.Age^2) + x.Gender, family = structure(list( #> family = "quasibinomial", link = "logit", linkfun = function (mu) #> .Call(C_logit_link, mu), linkinv = function (eta) #> .Call(C_logit_linkinv, eta), variance = function (mu) #> mu * (1 - mu), dev.resids = function (y, mu, wt) #> .Call(C_binomial_dev_resids, y, mu, wt), aic = function (y, #> n, mu, wt, dev) #> NA, mu.eta = function (eta) #> .Call(C_logit_mu_eta, eta), initialize = { #> if (NCOL(y) == 1) { #> if (is.factor(y)) #> y <- y != levels(y)[1L] #> n <- rep.int(1, nobs) #> y[weights == 0] <- 0 #> if (any(y < 0 | y > 1)) #> stop("y values must be 0 <= y <= 1") #> mustart <- (weights * y + 0.5)/(weights + 1) #> m <- weights * y #> if ("quasibinomial" == "binomial" && any(abs(m - #> round(m)) > 0.001)) #> warning(gettextf("non-integer #successes in a %s glm!", #> "quasibinomial"), domain = NA) #> } #> else if (NCOL(y) == 2) { #> if ("quasibinomial" == "binomial" && any(abs(y - #> round(y)) > 0.001)) #> warning(gettextf("non-integer counts in a %s glm!", #> "quasibinomial"), domain = NA) #> n <- (y1 <- y[, 1L]) + y[, 2L] #> y <- y1/n #> if (any(n0 <- n == 0)) #> y[n0] <- 0 #> weights <- weights * n #> mustart <- (n * y + 0.5)/(n + 1) #> } #> else stop(gettextf("for the '%s' family, y must be a vector of 0 and 1's\nor a 2 column matrix where col 1 is no. successes and col 2 is no. failures", #> "quasibinomial"), domain = NA) #> }, validmu = function (mu) #> all(is.finite(mu)) && all(0 < mu & mu < 1), valideta = function (eta) #> TRUE, dispersion = NA_real_), class = "family"), data = structure(list( #> x.Age = c(51, 50, 29, 28, 3, 56, 59, 42, 78, 47, 32, 37, #> 63, 37, 43, 72, 71, 1, 37, 46, 78, 61, 50, 44, 58, 48, 37, #> 59, 45, 47, 37, 34, 61, 78, 51, 45, 41, 51, 37, 52, 59, 37, #> 40, 34, 27, 49, 45, 45, 55, 52, 42, 51, 35, 52, 47, 47, 42, #> 25, 36, 54, 74, 49, 21, 93, 59, 51, 59, 42, 46, 48, 57, 78, #> 37, 51, 31, 34, 18, 37, 46, 49, 54, 45, 78, 48, 30, 78, 17, #> 61, 73, 53, 22, 16, 52, 64, 42, 33, 50, 55, 37, 35, 19, 30, #> 36, 51, 56, 37, 40, 37, 49, 49, 40, 67, 60, 37, 49, 33, 27, #> 26, 43, 78, 40, 51, 78, 37, 26, 48, 56, 78, 78, 31, 47, 36, #> 44, 43, 29, 44, 71, 37, 85, 26, 45, 44, 78, 15, 52, 41, 28, #> 48, 46, 37, 40, 37, 45, 23, 51, 45, 28, 46, 35, 55, 54, 37, #> 37, 45, 16, 64, 50, 32, 17, 58, 64, 49, 37, 37, 37, 37, 37, #> 27, 61, 45, 41, 58, 29, 19, 81, 42, 59, 34, 26, 58, 63, 27, #> 56, 58, 37, 54, 29, 37, 51, 55, 34, 37, 39, 37, 37, 26, 62, #> 61, 44, 37, 53, 41, 78, 34, 52, 37, 30, 37, 28, 37, 53, 24, #> 43, 37, 33, 51, 62, 52, 37, 45, 53, 52, 20, 70, 6, 57, 52, #> 55, 51, 70, 46, 45, 78, 69, 47, 41, 15, 67, 37, 33, 45, 36, #> 68, 28, 26, 37, 64, 47, 51, 57, 37, 20, 42, 53, 43, 37, 22, #> 44, 60, 40, 37, 27, 36, 37, 9, 79, 37, 48, 32, 25, 47, 3, #> 56, 33, 61, 39, 51, 60, 58, 13, 2, 36, 64, 42, 50, 35, 37, #> 78, 37, 36, 59, 42, 63, 68, 67, 80, 78, 58, 47, 37, 46, 45, #> 48, 51, 44, 38, 42, 27, 37, 53, 47, 37, 41, 57, 55, 46, 15, #> 51, 59, 71, 40, 57, 38, 23, 37, 73, 46, 37, 39, 37, 25, 34, #> 30, 47, 56, 28, 48, 43, 47, 35, 32, 43, 53, 39, 40, 43, 35, #> 37, 70, 35, 51, 7, 27, 43, 55, 44, 84, 43, 51, 37, 60, 39, #> 48, 51, 45, 52, 78, 37, 30, 29, 73, 43, 35, 37, 28, 54, 21, #> 27, 63, 36, 55, 52, 45, 41, 45, 59, 72, 45, 45, 37, 33.5, #> 50, 61, 41, 41, 41, 48, 11, 34, 42, 41, 41, 40, 50, 42, 43, #> 61, 35, 25, 41, 59, 17, 46, 50, 51, 41, 27, 41, 41, 37, 53, #> 36, 57, 30, 60, 50, 42, 47, 32, 40, 35, 46, 33.5, 68, 54, #> 41, 36, 29, 67, 37, 36, 59, 40, 50, 50, 41, 46, 33, 50, 45, #> 65, 50, 50, 64, 41, 63, 53, 21, 41, 40, 43, 33.5, 41, 37, #> 50, 33, 43, 16, 68, 41, 31, 55, 42, 42, 42, 55, 66, 44, 33.5, #> 43, 31, 68, 38, 51, 22, 45, 41, 41, 45, 79, 63, 45, 63, 47, #> 68, 41, 39, 5, 21, 60, 16, 67, 46, 54, 35, 41, 43, 46, 45, #> 44, 47, 53, 38, 43, 32, 25, 23, 51, 57, 50, 69, 63, 56, 53, #> 40, 19, 44, 39, 41, 41, 36, 33, 50, 43, 78, 33.5, 39, 43, #> 59, 41, 55, 41, 41, 26, 54, 38, 70, 60, 42, 48, 24, 33.5, #> 36, 47, 50, 68, 41, 54, 82, 33, 41, 54, 51, 59, 25, 33.5, #> 82, 52, 26, 22, 35, 74, 41, 41, 53, 29, 51, 53, 33.5, 40, #> 14, 40, 63, 50, 48, 45, 38, 22, 59, 65, 49, 78, 50, 25, 50, #> 60, 37, 50, 42, 45, 52, 50, 29, 33.5, 32, 58, 52, 68, 43, #> 52, 69, 32, 47, 50, 28, 50, 46, 18, 50, 50, 41, 33.5, 20, #> 61, 41, 40, 13, 28, 49, 27, 53, 62, 33.5, 37, 51, 40, 64, #> 82, 57, 45, 22, 33.5, 33.5, 72, 42, 55, 30, 39, 21, 40, 34, #> 33.5, 50, 11, 50, 21, 52, 34, 49, 48, 44, 25, 52, 26, 43, #> 73, 61, 34, 35, 67, 18, 44, 42, 43, 25, 41, 47, 60, 50, 42, #> 32, 27, 26, 56, 54, 24, 50, 44, 20, 41, 47, 47, 39, 38, 56, #> 40, 36, 60, 59, 28, 79, 35, 29, 47, 101, 69, 50, 63, 54, #> 56, 78, 39, 70, 37, 49, 50, 31, 37, 41, 55, 69, 40, 44, 37, #> 33, 31, 67, 24, 41, 41, 68, 37, 50, 51, 67, 42, 41, 50, 44, #> 31, 81, 52, 33.5, 29, 32, 51, 33, 33.5, 39, 71, 57, 57, 63, #> 62, 65, 34, 55, 34, 2, 18, 57, 43, 63, 57, 59, 35, 41, 50, #> 28, 46, 37, 33.5, 40, 52, 11, 33, 50, 50, 56, 48, 39, 50, #> 33, 50, 73, 39, 26, 48, 54, 69, 51, 50, 45, 57, 56, 48, 27, #> 33.5, 51, 23, 33.5, 66, 39, 40, 23, 45, 26, 84, 54, 49, 64, #> 47, 47, 13, 66, 20, 40, 41, 39, 50, 41, 69, 48, 59, 35, 51, #> 33.5, 35, 47, 54, 30, 56, 34, 41, 50, 76, 33.5, 47, 41, 44, #> 58, 59, 44, 33.5, 31, 40, 43, 56, 50, 33.5, 54, 58, 52, 50, #> 34, 41, 51, 38, 69, 53, 41, 31, 40, 34, 33.5, 49, 34, 41, #> 22, 43, 54, 51, 42, 57, 48, 40, 65, 33, 27, 50, 69, 77, 31, #> 45, 60, 20, 59, 50, 50, 32, 33.5, 55, 55, 56, 26, 67, 46, #> 33.5, 40, 33, 30, 41, 57, 73, 20, 40, 26, 33.5, 49, 33.5, #> 34, 65, 50, 41, 37, 66, 41, 50, 50, 30, 33, 41, 49, 48, 37, #> 64, 32, 42, 48, 22, 33.5, 35, 34, 24, 41, 51, 86, 33.5, 41, #> 35, 55, 50, 57, 8, 41, 41, 61, 23, 43, 54, 27, 50, 58, 31, #> 46, 43, 52, 41, 51, 43, 41, 50, 58, 39, 41, 33.5, 35, 23, #> 42, 50, 19, 33, 32, 64, 25, 36, 63, 20, 24, 34, 82, 36, 41, #> 33.5, 51, 43, 70, 39, 57, 31), x.Gender = structure(c(1L, #> 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, #> 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, #> 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, #> 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, #> 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, #> 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, #> 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, #> 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, #> 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, #> 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, #> 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, #> 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, #> 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, #> 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, #> 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, #> 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, #> 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, #> 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, #> 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, #> 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, #> 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, #> 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, #> 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, #> 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, #> 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, #> 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, #> 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 2L, 2L, 2L, 2L, 2L, 2L, #> 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, #> 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, #> 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, #> 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, #> 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, #> 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, #> 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, #> 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, #> 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, #> 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, #> 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, #> 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, #> 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, #> 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, #> 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, #> 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, #> 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, #> 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, #> 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, #> 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, #> 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, #> 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, #> 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, #> 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, #> 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, #> 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, #> 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, #> 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, #> 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, #> 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, #> 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, #> 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, #> 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, #> 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, #> 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, #> 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, #> 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, #> 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, #> 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, #> 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L), levels = c("0", "1"), class = "factor"), #> Smoke = structure(c(2L, 1L, 1L, 1L, 1L, 2L, 1L, 1L, 1L, 2L, #> 1L, 1L, 1L, 1L, 2L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, #> 1L, 1L, 1L, 2L, 2L, 1L, 1L, 1L, 1L, 2L, 2L, 1L, 2L, 1L, 2L, #> 2L, 1L, 1L, 1L, 1L, 2L, 2L, 2L, 2L, 1L, 2L, 2L, 1L, 2L, 1L, #> 2L, 2L, 1L, 1L, 2L, 1L, 2L, 1L, 1L, 1L, 2L, 1L, 1L, 1L, 2L, #> 1L, 1L, 1L, 2L, 1L, 1L, 1L, 1L, 2L, 2L, 2L, 1L, 1L, 1L, 1L, #> 1L, 1L, 1L, 1L, 2L, 1L, 1L, 2L, 1L, 2L, 1L, 1L, 2L, 1L, 1L, #> 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 2L, 2L, 1L, 1L, 2L, 1L, 1L, #> 1L, 1L, 1L, 1L, 1L, 1L, 2L, 1L, 1L, 1L, 1L, 2L, 1L, 1L, 1L, #> 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 2L, 2L, 1L, 1L, 1L, #> 2L, 1L, 2L, 2L, 1L, 1L, 1L, 2L, 1L, 2L, 2L, 1L, 2L, 1L, 2L, #> 1L, 1L, 1L, 2L, 1L, 1L, 1L, 1L, 1L, 2L, 1L, 2L, 1L, 1L, 1L, #> 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, #> 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 2L, 1L, 1L, 1L, 1L, 1L, 1L, #> 1L, 1L, 1L, 2L, 1L, 1L, 2L, 1L, 1L, 2L, 1L, 1L, 1L, 1L, 1L, #> 2L, 1L, 1L, 1L, 2L, 2L, 1L, 1L, 1L, 1L, 1L, 2L, 1L, 1L, 1L, #> 1L, 2L, 1L, 2L, 1L, 1L, 1L, 1L, 1L, 2L, 1L, 1L, 1L, 1L, 1L, #> 2L, 1L, 1L, 1L, 1L, 1L, 1L, 2L, 2L, 1L, 1L, 1L, 1L, 1L, 1L, #> 1L, 1L, 2L, 2L, 1L, 1L, 2L, 2L, 1L, 1L, 1L, 1L, 2L, 1L, 1L, #> 1L, 1L, 1L, 1L, 2L, 1L, 2L, 1L, 2L, 1L, 1L, 2L, 2L, 1L, 1L, #> 1L, 1L, 1L, 1L, 1L, 1L, 2L, 1L, 1L, 1L, 1L, 1L, 2L, 2L, 1L, #> 2L, 2L, 1L, 2L, 2L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 2L, 1L, 2L, #> 2L, 1L, 2L, 1L, 1L, 1L, 2L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, #> 1L, 2L, 1L, 2L, 1L, 1L, 1L, 2L, 2L, 1L, 1L, 1L, 1L, 2L, 1L, #> 1L, 1L, 1L, 1L, 1L, 2L, 1L, 1L, 2L, 2L, 1L, 1L, 2L, 2L, 1L, #> 1L, 1L, 1L, 2L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, #> 1L, 1L, 1L, 2L, 1L, 1L, 2L, 1L, 1L, 2L, 2L, 1L, 2L, 2L, 1L, #> 1L, 2L, 1L, 2L, 2L, 2L, 2L, 1L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, #> 2L, 1L, 2L, 1L, 2L, 2L, 1L, 2L, 2L, 1L, 2L, 1L, 2L, 2L, 1L, #> 1L, 2L, 2L, 1L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 1L, 1L, 1L, #> 2L, 1L, 2L, 1L, 2L, 2L, 2L, 2L, 2L, 2L, 1L, 2L, 1L, 2L, 2L, #> 1L, 2L, 2L, 2L, 2L, 2L, 2L, 1L, 2L, 1L, 2L, 1L, 2L, 1L, 2L, #> 1L, 2L, 1L, 1L, 2L, 2L, 2L, 2L, 2L, 1L, 2L, 2L, 2L, 1L, 2L, #> 1L, 2L, 2L, 2L, 1L, 1L, 2L, 2L, 2L, 1L, 2L, 2L, 2L, 2L, 1L, #> 2L, 2L, 1L, 1L, 2L, 1L, 2L, 2L, 2L, 1L, 2L, 2L, 2L, 2L, 2L, #> 2L, 2L, 2L, 2L, 1L, 1L, 1L, 2L, 1L, 2L, 1L, 1L, 2L, 2L, 2L, #> 1L, 2L, 1L, 2L, 2L, 1L, 2L, 2L, 2L, 1L, 1L, 2L, 2L, 1L, 2L, #> 2L, 2L, 2L, 1L, 2L, 2L, 1L, 2L, 2L, 2L, 1L, 1L, 1L, 2L, 2L, #> 1L, 2L, 2L, 1L, 1L, 2L, 1L, 2L, 2L, 1L, 1L, 1L, 2L, 1L, 1L, #> 1L, 1L, 2L, 2L, 1L, 2L, 2L, 2L, 1L, 2L, 1L, 2L, 1L, 2L, 2L, #> 2L, 2L, 1L, 1L, 2L, 2L, 1L, 2L, 1L, 2L, 1L, 2L, 2L, 2L, 2L, #> 2L, 2L, 2L, 1L, 1L, 1L, 1L, 1L, 1L, 2L, 1L, 1L, 2L, 2L, 2L, #> 2L, 2L, 1L, 2L, 2L, 2L, 1L, 1L, 1L, 2L, 2L, 1L, 1L, 2L, 1L, #> 2L, 1L, 1L, 2L, 2L, 2L, 1L, 1L, 2L, 2L, 1L, 1L, 1L, 1L, 2L, #> 1L, 1L, 2L, 1L, 2L, 1L, 1L, 2L, 1L, 2L, 1L, 1L, 2L, 2L, 2L, #> 2L, 1L, 2L, 1L, 2L, 1L, 1L, 2L, 2L, 1L, 1L, 2L, 2L, 1L, 1L, #> 2L, 2L, 2L, 2L, 2L, 2L, 1L, 1L, 2L, 2L, 1L, 2L, 2L, 1L, 2L, #> 2L, 2L, 2L, 1L, 2L, 2L, 2L, 2L, 2L, 1L, 1L, 2L, 1L, 2L, 1L, #> 1L, 2L, 2L, 1L, 2L, 1L, 2L, 1L, 1L, 2L, 2L, 1L, 2L, 2L, 2L, #> 1L, 1L, 2L, 1L, 2L, 1L, 1L, 1L, 2L, 2L, 1L, 2L, 2L, 2L, 1L, #> 2L, 2L, 2L, 2L, 1L, 1L, 2L, 1L, 2L, 1L, 2L, 1L, 1L, 2L, 1L, #> 2L, 2L, 1L, 2L, 1L, 2L, 2L, 2L, 1L, 1L, 1L, 2L, 1L, 1L, 2L, #> 2L, 2L, 2L, 1L, 2L, 2L, 1L, 2L, 2L, 1L, 1L, 2L, 2L, 2L, 1L, #> 1L, 2L, 2L, 2L, 1L, 2L, 1L, 2L, 2L, 1L, 2L, 2L, 2L, 2L, 2L, #> 1L, 1L, 1L, 2L, 1L, 1L, 1L, 2L, 1L, 1L, 2L, 1L, 1L, 2L, 2L, #> 1L, 2L, 2L, 1L, 1L, 1L, 2L, 2L, 2L, 2L, 2L, 1L, 2L, 2L, 1L, #> 2L, 1L, 1L, 2L, 2L, 1L, 2L, 1L, 2L, 2L, 1L, 1L, 2L, 2L, 2L, #> 2L, 1L, 1L, 1L, 1L, 2L, 1L, 2L, 2L, 1L, 2L, 2L, 2L, 2L, 1L, #> 2L, 2L, 2L, 1L, 2L, 2L, 2L, 2L, 1L, 1L, 2L, 2L, 2L, 1L, 2L, #> 2L, 1L, 2L, 2L, 2L, 1L, 1L, 2L, 1L, 2L, 1L, 1L, 1L, 2L, 1L, #> 1L, 2L, 2L, 2L, 1L, 1L, 2L, 1L, 2L, 1L, 1L, 2L, 1L, 1L, 2L, #> 2L, 2L, 1L, 1L, 1L, 1L, 1L, 1L, 2L, 1L, 1L, 2L, 2L, 2L, 1L, #> 1L, 2L, 2L, 2L, 1L, 1L, 2L, 2L, 2L, 1L, 2L, 1L, 1L, 2L, 1L, #> 1L, 2L, 2L, 1L, 2L, 2L, 1L, 1L, 2L, 2L, 2L, 2L, 1L, 1L, 2L, #> 1L, 1L, 1L, 2L, 2L, 1L, 2L, 2L, 2L, 1L, 2L, 2L, 2L, 2L, 2L, #> 2L, 2L, 1L, 2L, 2L, 1L, 1L, 2L, 2L, 2L, 1L, 1L, 1L, 1L, 1L, #> 1L, 2L, 1L, 1L, 2L, 1L, 2L, 2L, 1L, 2L, 2L, 1L, 2L, 2L, 2L #> ), levels = c("0", "1"), class = "factor"), CVD = structure(c(2L, #> 1L, 1L, 1L, 1L, 2L, 1L, 1L, 2L, 1L, 1L, 1L, 1L, 1L, 2L, 1L, #> 2L, 1L, 1L, 1L, 2L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 2L, 1L, #> 1L, 1L, 2L, 2L, 1L, 1L, 2L, 1L, 2L, 2L, 1L, 1L, 1L, 1L, 2L, #> 1L, 1L, 2L, 1L, 2L, 2L, 1L, 2L, 1L, 2L, 1L, 1L, 1L, 2L, 2L, #> 2L, 1L, 2L, 1L, 2L, 1L, 1L, 1L, 1L, 1L, 2L, 1L, 2L, 1L, 1L, #> 1L, 1L, 1L, 2L, 2L, 1L, 2L, 1L, 1L, 2L, 1L, 1L, 1L, 2L, 1L, #> 1L, 2L, 1L, 1L, 1L, 1L, 2L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, #> 1L, 1L, 2L, 2L, 1L, 2L, 2L, 1L, 1L, 1L, 1L, 1L, 1L, 2L, 1L, #> 2L, 2L, 1L, 1L, 1L, 2L, 2L, 2L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, #> 1L, 1L, 2L, 1L, 1L, 1L, 2L, 1L, 1L, 1L, 1L, 2L, 1L, 1L, 1L, #> 1L, 1L, 1L, 2L, 1L, 1L, 2L, 1L, 2L, 1L, 1L, 1L, 1L, 1L, 1L, #> 1L, 1L, 1L, 2L, 1L, 2L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, #> 1L, 1L, 1L, 2L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, #> 1L, 1L, 2L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, #> 1L, 2L, 1L, 2L, 1L, 1L, 1L, 1L, 1L, 2L, 1L, 1L, 1L, 1L, 2L, #> 1L, 1L, 1L, 1L, 1L, 2L, 1L, 2L, 1L, 1L, 2L, 1L, 2L, 1L, 1L, #> 1L, 2L, 2L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 2L, 1L, 1L, 1L, #> 2L, 1L, 2L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 2L, 1L, 1L, #> 1L, 1L, 1L, 1L, 2L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 2L, 1L, #> 2L, 1L, 2L, 1L, 1L, 1L, 2L, 1L, 1L, 1L, 1L, 2L, 1L, 1L, 1L, #> 2L, 1L, 1L, 1L, 2L, 2L, 2L, 1L, 1L, 1L, 1L, 1L, 2L, 1L, 1L, #> 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 2L, 2L, 1L, 2L, 1L, 1L, 1L, #> 2L, 1L, 1L, 1L, 2L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 2L, 1L, 1L, #> 1L, 1L, 2L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 2L, 1L, 2L, #> 1L, 1L, 1L, 2L, 1L, 2L, 2L, 2L, 1L, 1L, 1L, 1L, 2L, 1L, 1L, #> 2L, 1L, 1L, 1L, 2L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 2L, 1L, 1L, #> 2L, 1L, 1L, 1L, 2L, 2L, 1L, 1L, 1L, 1L, 2L, 1L, 1L, 1L, 1L, #> 1L, 1L, 1L, 1L, 1L, 1L, 1L, 2L, 1L, 2L, 1L, 1L, 1L, 2L, 2L, #> 1L, 1L, 2L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 2L, 1L, 2L, 2L, #> 2L, 2L, 1L, 1L, 1L, 1L, 1L, 2L, 1L, 1L, 1L, 1L, 2L, 1L, 1L, #> 2L, 1L, 2L, 2L, 1L, 1L, 1L, 2L, 2L, 1L, 2L, 2L, 2L, 1L, 2L, #> 2L, 1L, 1L, 1L, 2L, 1L, 1L, 1L, 2L, 1L, 1L, 1L, 2L, 1L, 1L, #> 2L, 1L, 1L, 1L, 2L, 2L, 1L, 1L, 1L, 1L, 2L, 1L, 2L, 1L, 1L, #> 1L, 1L, 1L, 2L, 2L, 2L, 2L, 2L, 2L, 1L, 1L, 1L, 1L, 2L, 1L, #> 2L, 1L, 2L, 1L, 1L, 1L, 1L, 2L, 1L, 2L, 2L, 1L, 2L, 1L, 1L, #> 1L, 2L, 1L, 2L, 1L, 1L, 2L, 2L, 2L, 1L, 1L, 1L, 1L, 1L, 1L, #> 1L, 1L, 1L, 2L, 1L, 2L, 1L, 1L, 1L, 2L, 1L, 1L, 1L, 2L, 1L, #> 2L, 2L, 1L, 2L, 1L, 1L, 1L, 2L, 2L, 2L, 1L, 2L, 2L, 1L, 1L, #> 1L, 2L, 2L, 1L, 1L, 2L, 2L, 1L, 1L, 1L, 2L, 1L, 1L, 1L, 1L, #> 2L, 2L, 1L, 1L, 1L, 2L, 1L, 2L, 2L, 1L, 1L, 1L, 1L, 2L, 2L, #> 2L, 2L, 1L, 2L, 1L, 1L, 2L, 1L, 2L, 2L, 2L, 1L, 1L, 1L, 1L, #> 1L, 2L, 1L, 2L, 2L, 1L, 1L, 2L, 1L, 2L, 2L, 1L, 2L, 2L, 1L, #> 1L, 1L, 1L, 2L, 1L, 1L, 1L, 1L, 1L, 2L, 1L, 1L, 1L, 2L, 1L, #> 1L, 2L, 2L, 1L, 1L, 1L, 1L, 2L, 1L, 1L, 1L, 2L, 1L, 1L, 1L, #> 1L, 2L, 1L, 2L, 1L, 1L, 1L, 2L, 2L, 2L, 1L, 2L, 1L, 1L, 2L, #> 1L, 1L, 1L, 2L, 1L, 1L, 1L, 1L, 1L, 1L, 2L, 2L, 2L, 1L, 1L, #> 1L, 1L, 2L, 2L, 1L, 2L, 2L, 1L, 1L, 1L, 2L, 2L, 1L, 2L, 1L, #> 1L, 2L, 2L, 1L, 2L, 1L, 1L, 1L, 2L, 2L, 2L, 2L, 1L, 2L, 2L, #> 1L, 2L, 1L, 2L, 2L, 1L, 1L, 1L, 2L, 2L, 1L, 1L, 1L, 1L, 1L, #> 1L, 1L, 1L, 1L, 1L, 1L, 2L, 2L, 2L, 1L, 1L, 2L, 1L, 1L, 2L, #> 2L, 1L, 1L, 1L, 2L, 1L, 1L, 2L, 2L, 2L, 2L, 1L, 2L, 2L, 1L, #> 2L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 2L, 1L, 1L, 2L, 1L, 1L, 1L, #> 1L, 2L, 2L, 1L, 1L, 2L, 2L, 2L, 1L, 1L, 2L, 1L, 2L, 2L, 1L, #> 1L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 1L, 1L, 1L, 2L, 1L, 1L, #> 1L, 1L, 1L, 1L, 2L, 1L, 2L, 2L, 2L, 1L, 2L, 1L, 1L, 1L, 1L, #> 1L, 2L, 1L, 1L, 1L, 2L, 2L, 2L, 1L, 2L, 1L, 1L, 2L, 2L, 1L, #> 2L, 1L, 2L, 2L, 2L, 1L, 2L, 1L, 2L, 2L, 1L, 1L, 1L, 1L, 1L, #> 1L, 2L, 2L, 1L, 2L, 2L, 2L, 2L, 1L, 1L, 2L, 1L, 2L, 2L, 2L, #> 1L, 1L, 1L, 1L, 1L, 2L, 1L, 1L, 1L, 2L, 1L, 1L, 2L, 1L, 1L, #> 2L, 1L, 1L, 2L, 2L, 2L, 1L, 1L, 1L, 1L, 2L, 2L, 2L, 1L, 1L, #> 2L, 1L, 2L, 1L, 2L, 1L, 1L, 1L, 1L, 2L, 2L, 1L, 2L, 1L, 1L, #> 1L, 1L, 2L, 1L, 1L, 2L, 2L, 1L, 1L, 2L, 1L, 2L, 2L, 1L, 1L, #> 1L, 1L, 2L, 1L, 2L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 2L, #> 2L, 1L, 1L, 1L, 2L, 2L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 2L, 1L, #> 2L, 2L, 1L, 1L, 1L, 2L, 1L, 2L, 1L, 1L, 2L, 1L, 1L, 1L, 1L, #> 1L, 1L, 1L, 2L, 1L, 1L, 1L, 1L, 1L, 1L, 2L, 1L, 1L, 1L, 2L, #> 1L, 1L, 1L, 2L, 1L, 2L, 1L, 2L, 1L), levels = c("0", "1"), class = "factor")), row.names = c(NA, #> -1000L), class = "data.frame")) #> #> Coefficients: #> (Intercept) x.Age I(x.Age^2) x.Gender1 #> -28.99771 1.19555 -0.01223 2.12637 #> #> Degrees of Freedom: 999 Total (i.e. Null); 996 Residual #> Null Deviance: 1378 #> Residual Deviance: 819.7 AIC: NA