探索性分析
Show the code data ( lalonde , package= 'Matching' ) lalonde <- lalonde %>% mutate ( across ( treat , as.factor ) ) str ( lalonde ) #> 'data.frame': 445 obs. of 12 variables: #> $ age : int 37 22 30 27 33 22 23 32 22 33 ... #> $ educ : int 11 9 12 11 8 9 12 11 16 12 ... #> $ black : int 1 0 1 1 1 1 1 1 1 0 ... #> $ hisp : int 0 1 0 0 0 0 0 0 0 0 ... #> $ married: int 1 0 0 0 0 0 0 0 0 1 ... #> $ nodegr : int 1 1 0 1 1 1 0 1 0 0 ... #> $ re74 : num 0 0 0 0 0 0 0 0 0 0 ... #> $ re75 : num 0 0 0 0 0 0 0 0 0 0 ... #> $ re78 : num 9930 3596 24910 7506 290 ... #> $ u74 : int 1 1 1 1 1 1 1 1 1 1 ... #> $ u75 : int 1 1 1 1 1 1 1 1 1 1 ... #> $ treat : Factor w/ 2 levels "0","1": 2 2 2 2 2 2 2 2 2 2 ...
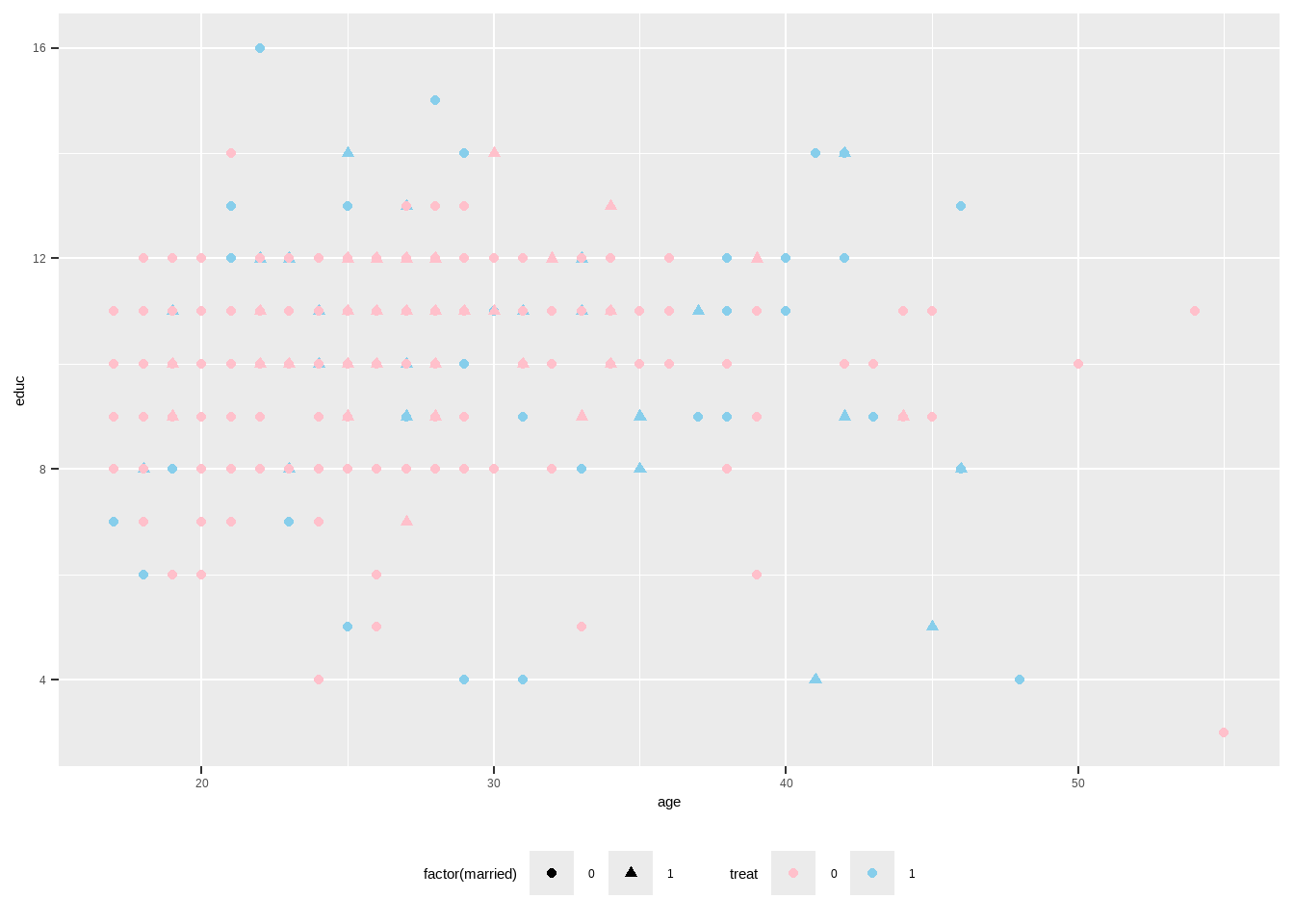
Show the code ggplot ( lalonde , aes ( x = age , y = educ , shape = factor ( married ) , color = treat ) ) + geom_point ( ) +
scale_color_manual ( values = c ( `0`= "pink" ,`1`= "skyblue" ) ) +
theme ( legend.position = 'bottom' )
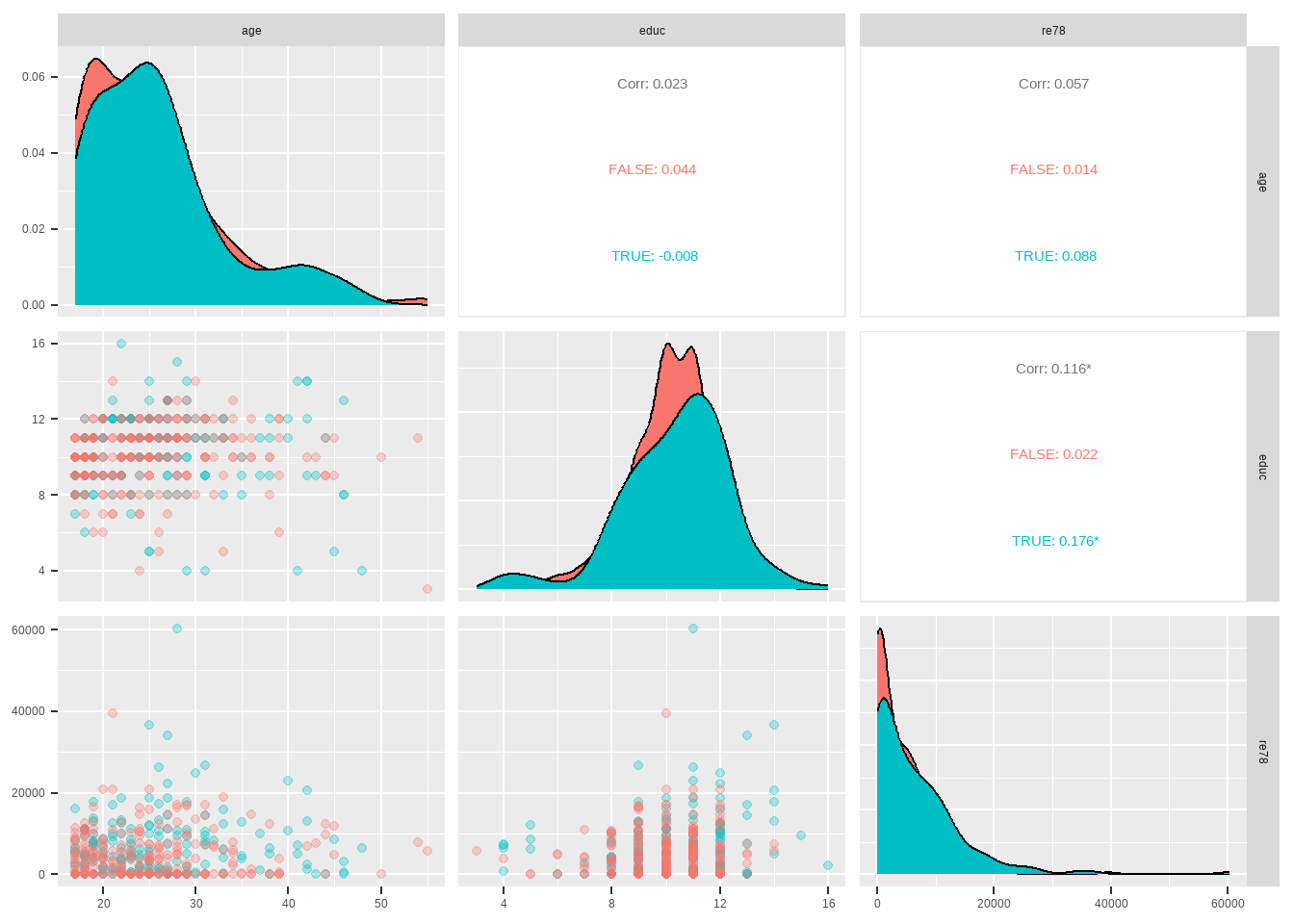
Show the code GGally :: ggpairs ( lalonde %>% select ( age ,educ , re78 , treat ) ,
columns = c ( 1 ,2 ,3 ) ,
mapping = ggplot2 :: aes ( color = treat == 1 ) ,
upper = list ( continuous = 'cor' ) ,
lower = list ( continuous = GGally :: wrap ( "points" , alpha = 0.3 ) ,
combo = GGally :: wrap ( "dot_no_facet" , alpha = 0.4 ) ) ,
diag = list ( continuous = "densityDiag" , discrete = "barDiag" , na = "naDiag" , alpha = 0.4 ) ,
)
估计倾向得分(逻辑回归)
Show the code lalonde_formula <- treat ~ age + I ( age ^ 2 ) + educ + I ( educ ^ 2 ) + black + hisp + married + nodegr + re74 + I ( re74 ^ 2 ) + re75 + I ( re75 ^ 2 )
logit_fit <- glm ( formula = lalonde_formula , data = lalonde ,
family = binomial ( link = 'logit' ) )
倾向性得分就是模型的拟合值,
Show the code lalonde $ ps <- fitted ( logit_fit )
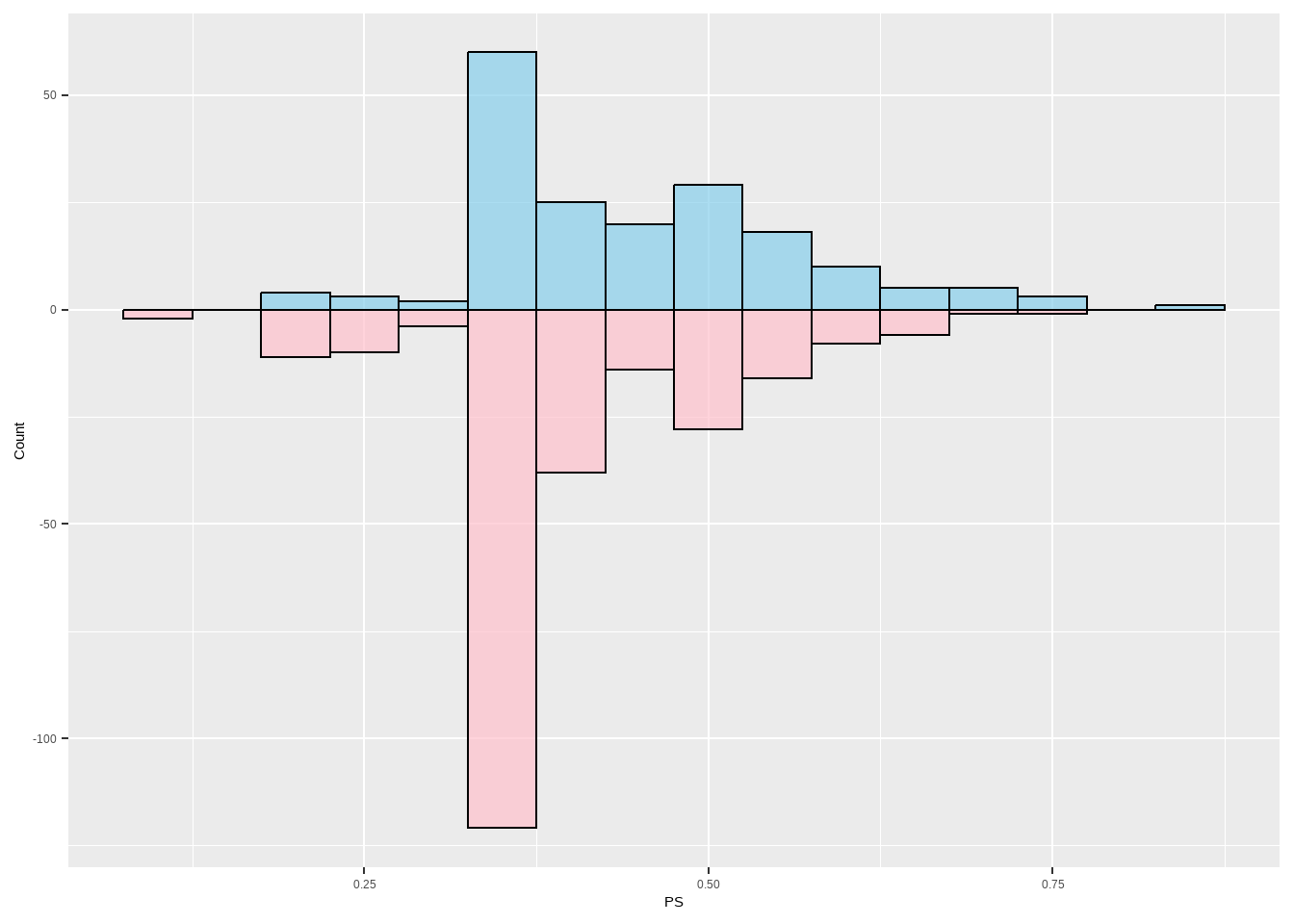
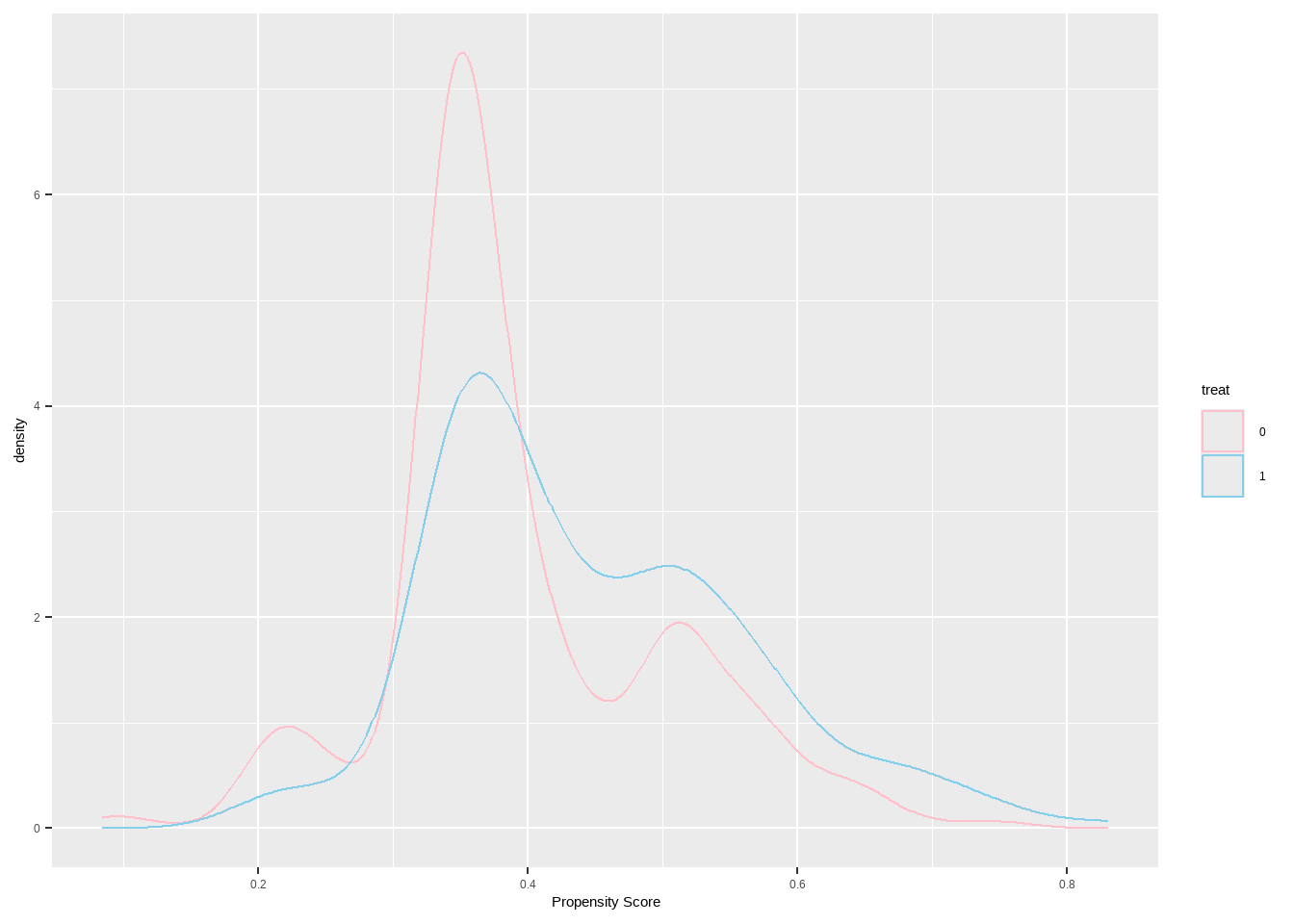
检查倾向得分的分布,以确保我们有良好的重叠
Show the code ggplot ( ) + geom_histogram ( data = subset ( lalonde , treat == 1 ) , aes ( x = ps , y = after_stat ( count ) ) ,
binwidth = 0.05 , fill = "skyblue" , color = "black" , alpha = 0.7 ) +
geom_histogram ( data = subset ( lalonde , treat == 0 ) , aes ( x = ps , y = - after_stat ( count ) ) ,
binwidth = 0.05 , fill = "pink" , color = "black" , alpha = 0.7 ) +
labs ( y = "Count" , x = "PS" )
Show the code ggplot ( lalonde , aes ( x = ps , color = treat ) ) + geom_density ( ) +
xlab ( 'Propensity Score' ) +
scale_color_manual ( values = c ( `0`= "pink" ,`1`= "skyblue" ) )
倾向得分分层
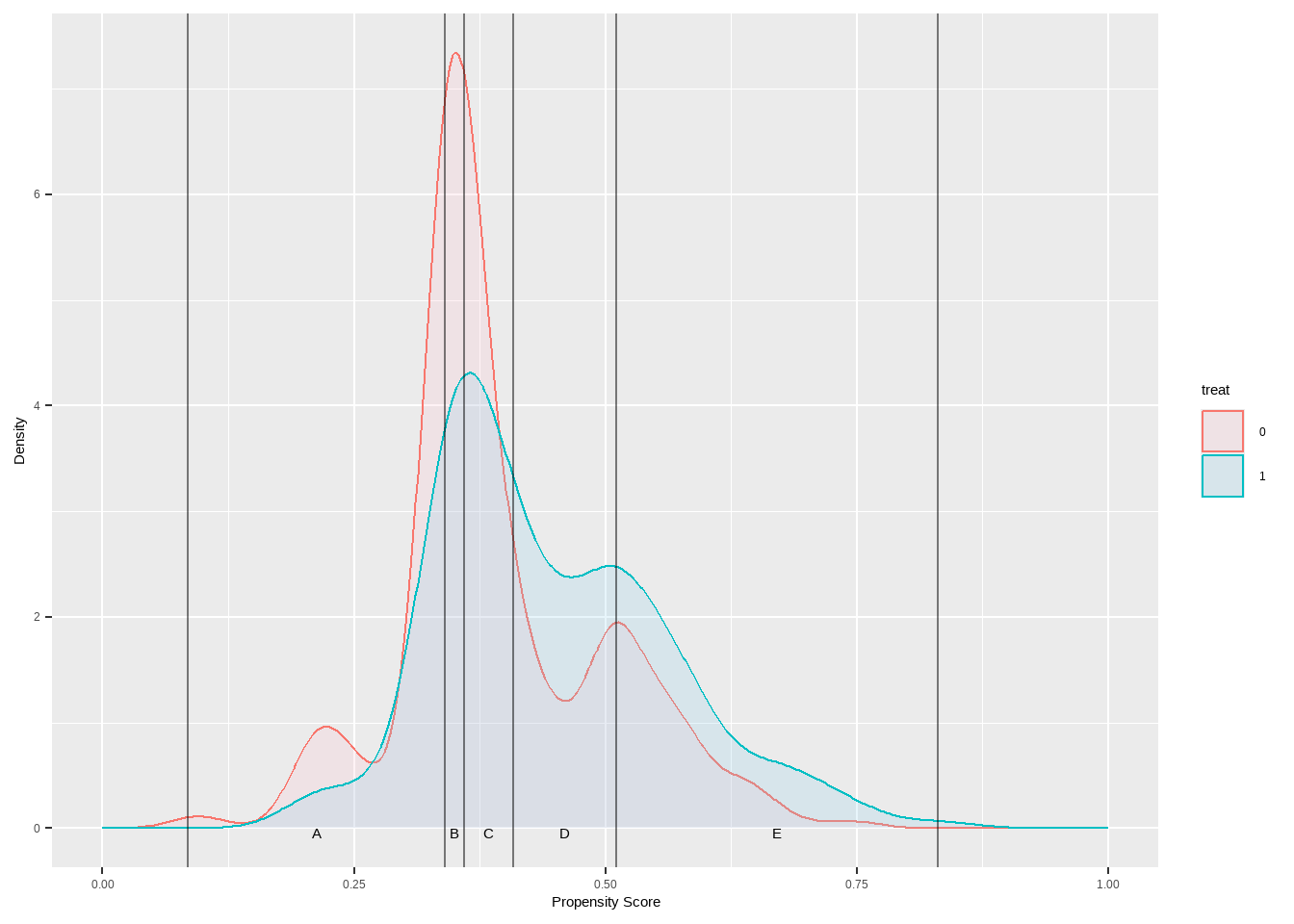
根据倾向分数使用五分位数进行分层
Show the code breaks5 <- psa :: get_strata_breaks ( lalonde $ ps ) str ( breaks5 ) #> List of 2 #> $ breaks: Named num [1:6] 0.0849 0.3403 0.3594 0.408 0.5112 ... #> ..- attr(*, "names")= chr [1:6] "0%" "20%" "40%" "60%" ... #> $ labels:'data.frame': 5 obs. of 4 variables: #> ..$ strata: chr [1:5] "A" "B" "C" "D" ... #> ..$ xmin : num [1:5] 0.0849 0.3403 0.3594 0.408 0.5112 #> ..$ xmax : num [1:5] 0.34 0.359 0.408 0.511 0.83 #> ..$ xmid : num [1:5] 0.213 0.35 0.384 0.46 0.671 lalonde $ lr_strata5 <- cut ( x = lalonde $ ps , breaks = breaks5 $ breaks ,
include.lowest = TRUE ,
labels = breaks5 $ labels $ strata )
table ( lalonde $ treat , lalonde $ lr_strata5 ) #> #> A B C D E #> 0 66 61 51 42 40 #> 1 23 28 38 47 49
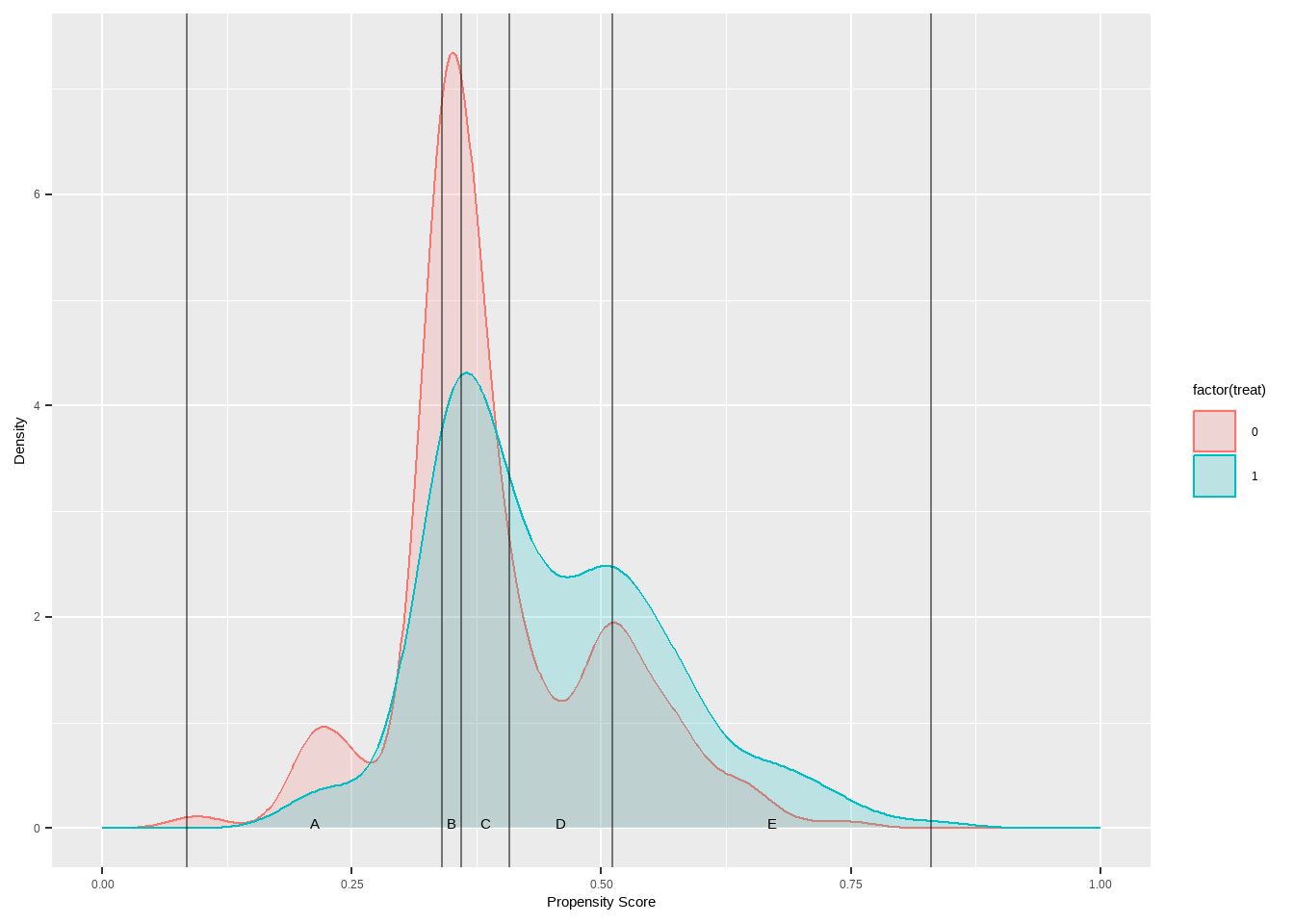
Show the code ggplot ( lalonde , aes ( x = ps , color = treat ) ) + geom_density ( aes ( fill = treat ) , alpha = 0.2 ) +
geom_vline ( xintercept = breaks5 $ breaks , alpha = 0.5 ) +
geom_text ( data = breaks5 $ labels ,
aes ( x = xmid , y = 0 , label = strata ) ,
color = 'black' , vjust = 1 ) +
xlab ( 'Propensity Score' ) + ylab ( 'Density' ) +
xlim ( c ( 0 , 1 ) ) +
scale_fill_manual ( values = c ( `0`= "pink" ,`1`= "skyblue" ) )
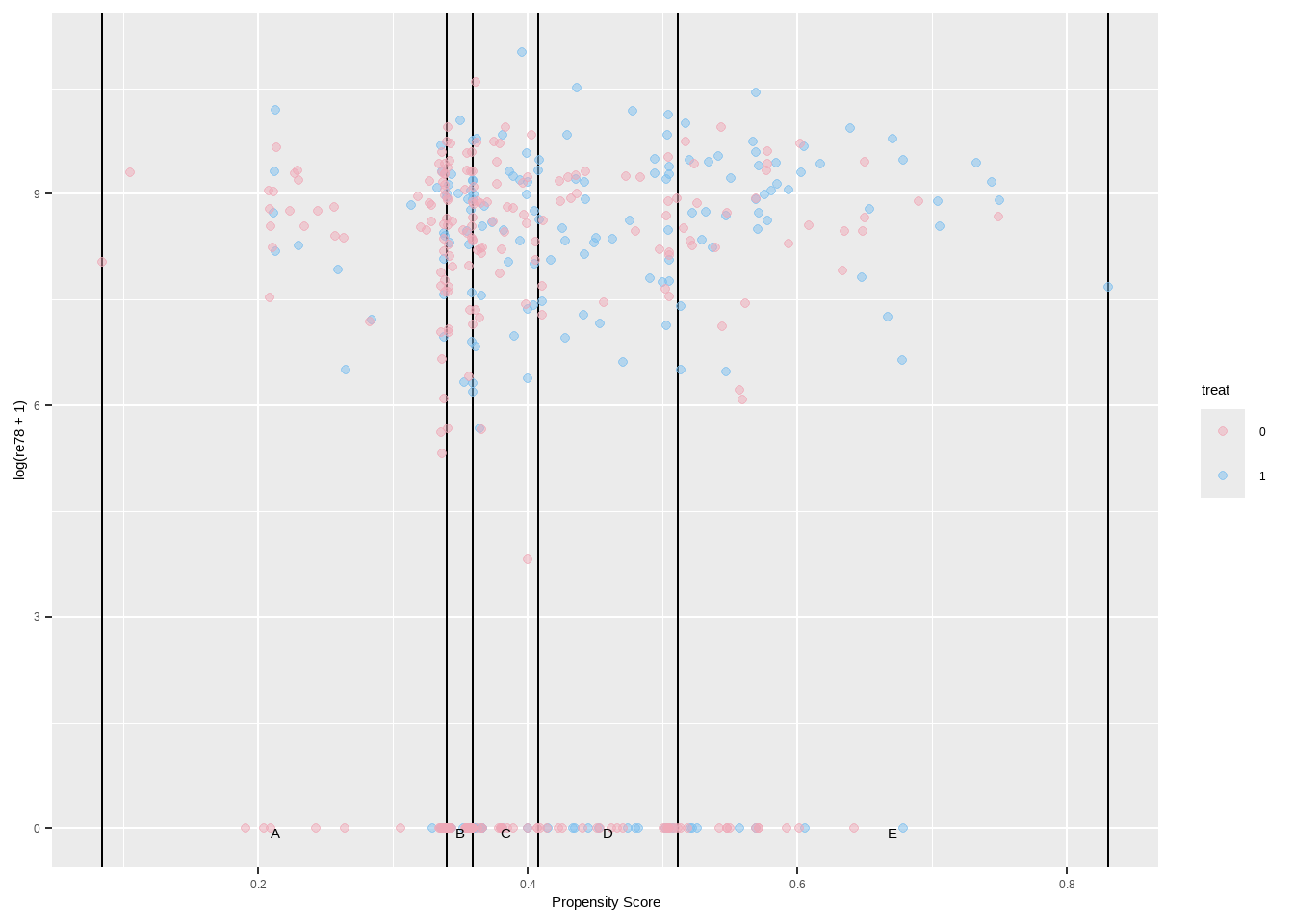
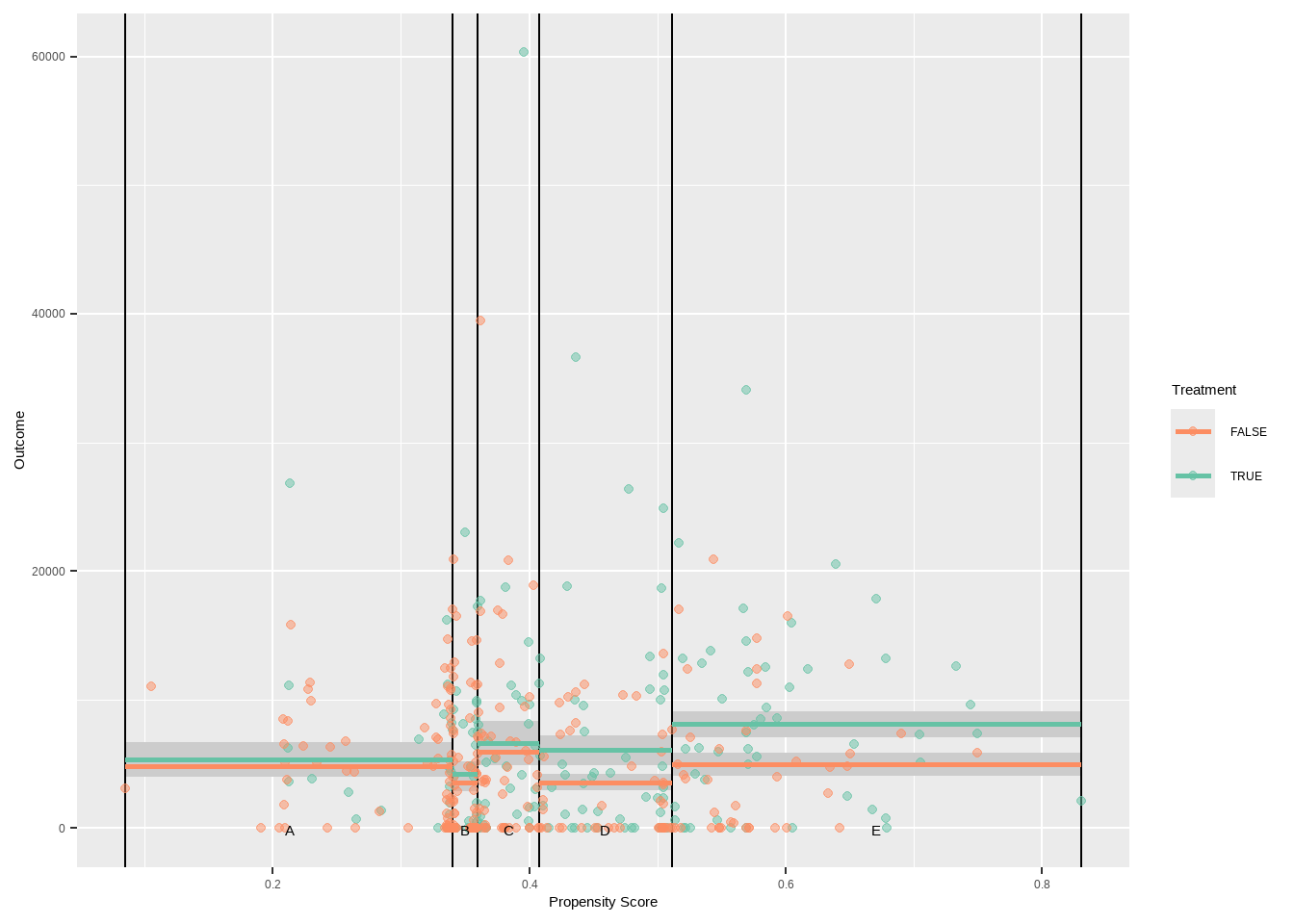
Show the code ggplot ( ) + geom_vline ( xintercept = breaks5 $ breaks ) +
geom_point ( data = lalonde , aes ( x = ps , y = log ( re78 + 1 ) , color = treat ) , alpha = 0.5 ) +
geom_text ( data = breaks5 $ labels , aes ( x = xmid , y = 0 , label = strata ) , color = 'black' , vjust = 1 ) +
xlab ( 'Propensity Score' ) +
scale_color_manual ( values = c ( `0`= "pink2" ,`1`= "skyblue2" ) )
平衡诊断
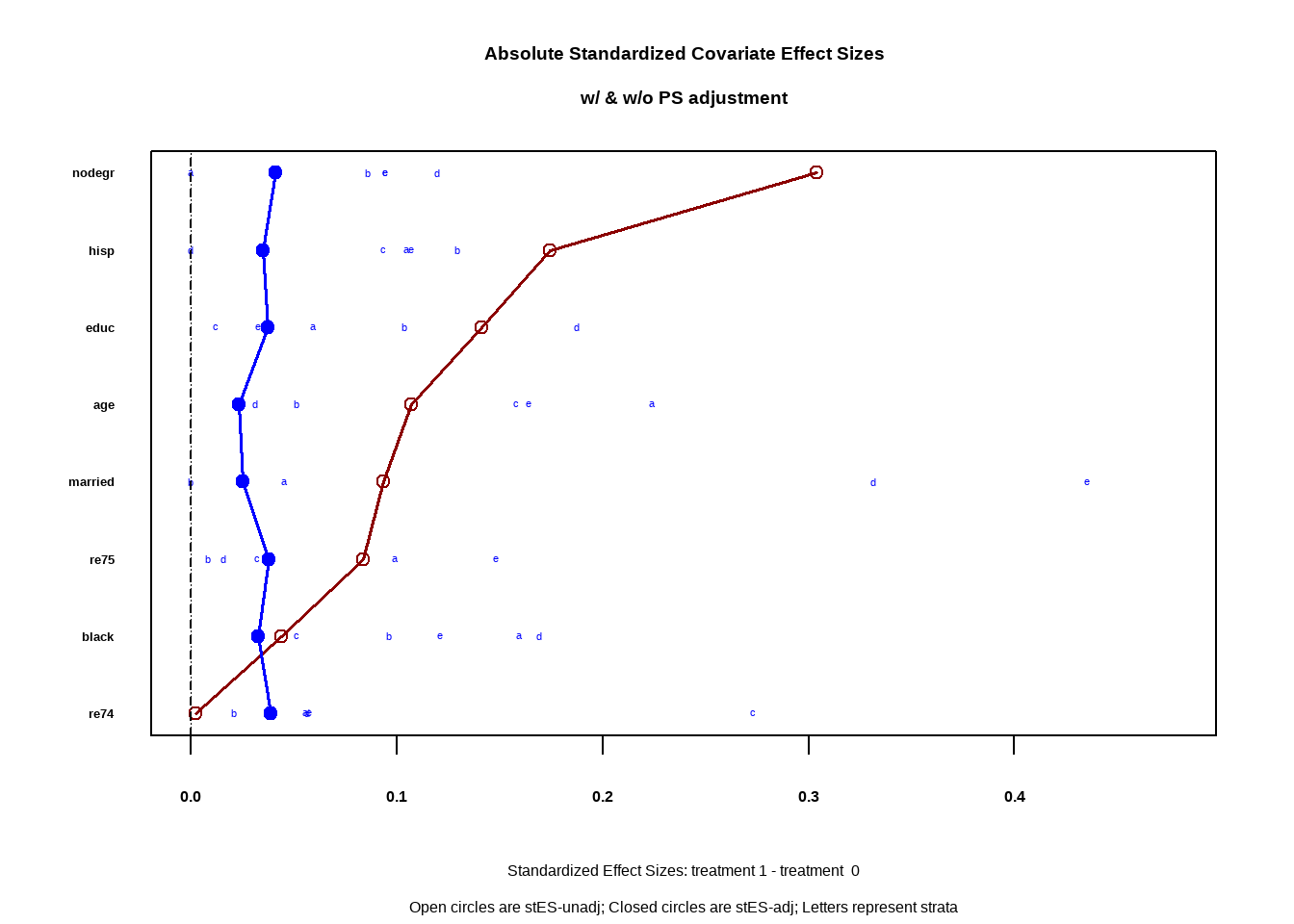
效应大小
Show the code covars <- all.vars ( lalonde_formula ) covars <- lalonde [ ,covars [ - 1 ] ] PSAgraphics :: cv.bal.psa ( covariates = covars , treatment = lalonde $ treat ,
propensity = lalonde $ ps ,
strata = lalonde $ lr_strata )
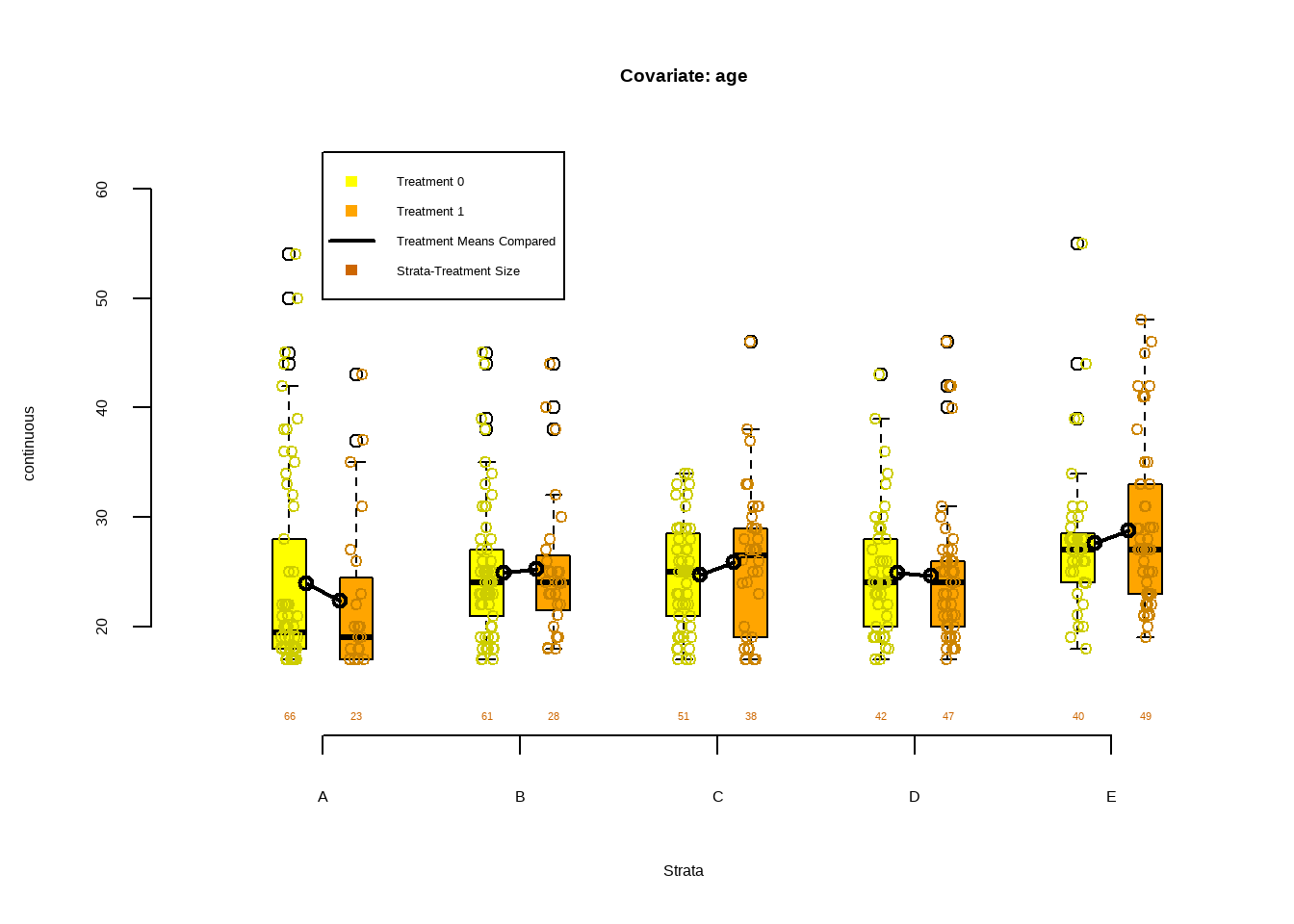
数值型协变量平衡图
Show the code PSAgraphics :: box.psa ( continuous = lalonde $ age , treatment = lalonde $ treat ,
strata = lalonde $ lr_strata ,
xlab = "Strata" ,
balance = FALSE ,
main = 'Covariate: age' )
分类型协变量平衡图
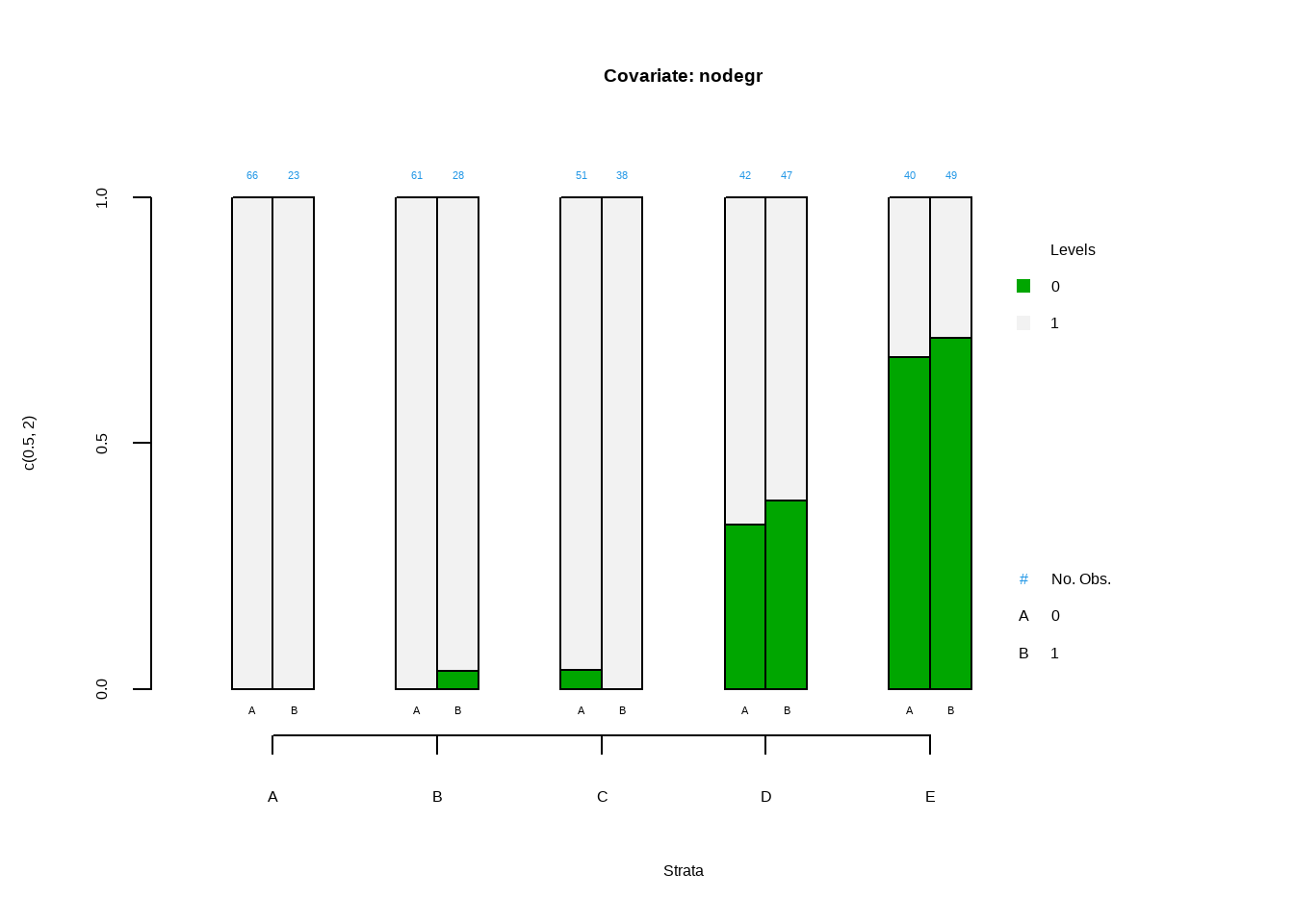
Show the code PSAgraphics :: cat.psa ( categorical = lalonde $ nodegr , treatment = lalonde $ treat ,
strata = lalonde $ lr_strata ,
xlab = 'Strata' ,
balance = FALSE ,
main = 'Covariate: nodegr' ) #> $`treatment:stratum.proportions`
#> 0:A 1:A 0:B 1:B 0:C 1:C 0:D 1:D 0:E 1:E
#> 0 0 0 0 0.036 0.039 0 0.333 0.383 0.675 0.714
#> 1 1 1 1 0.964 0.961 1 0.667 0.617 0.325 0.286
估计因果效应
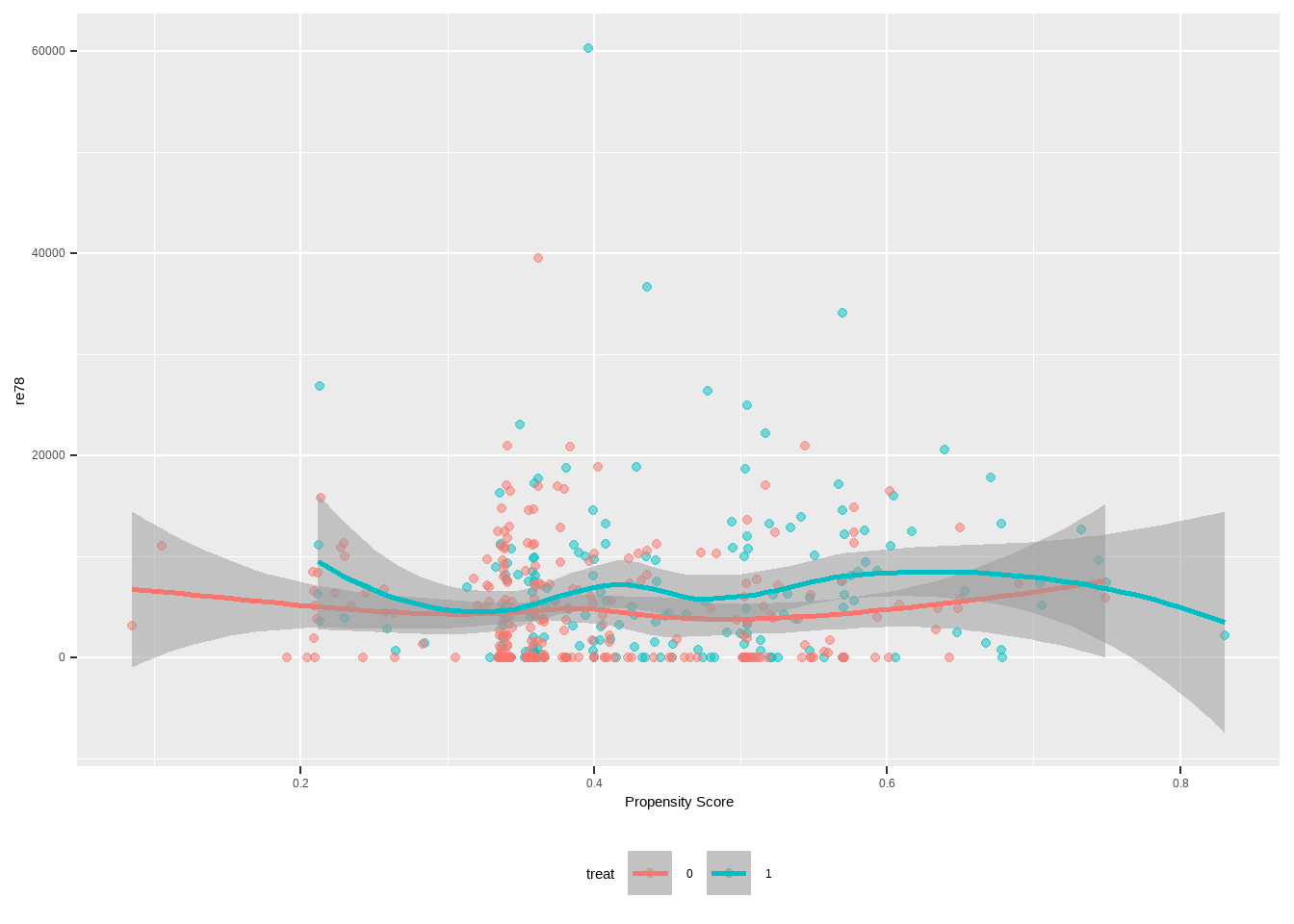
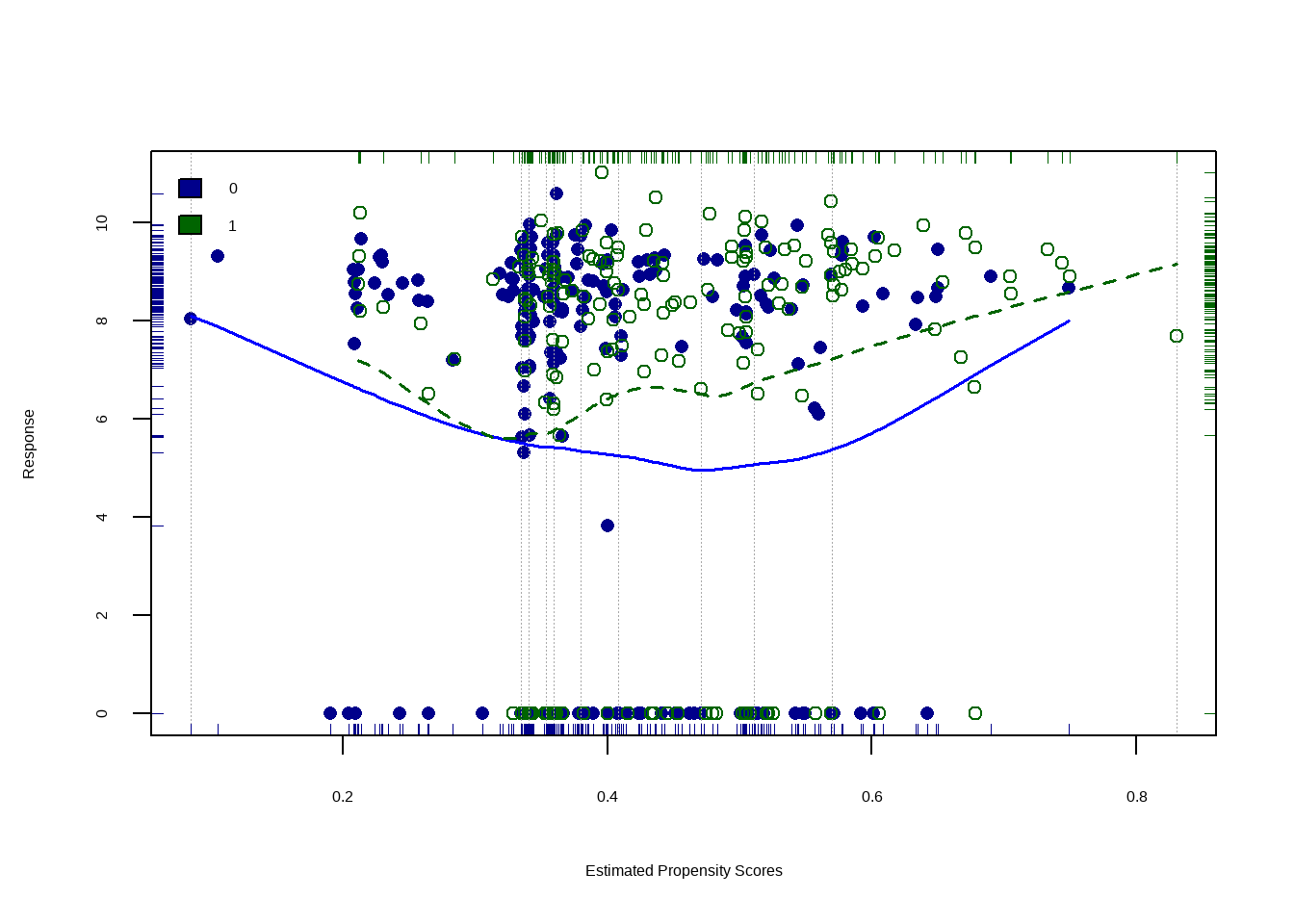
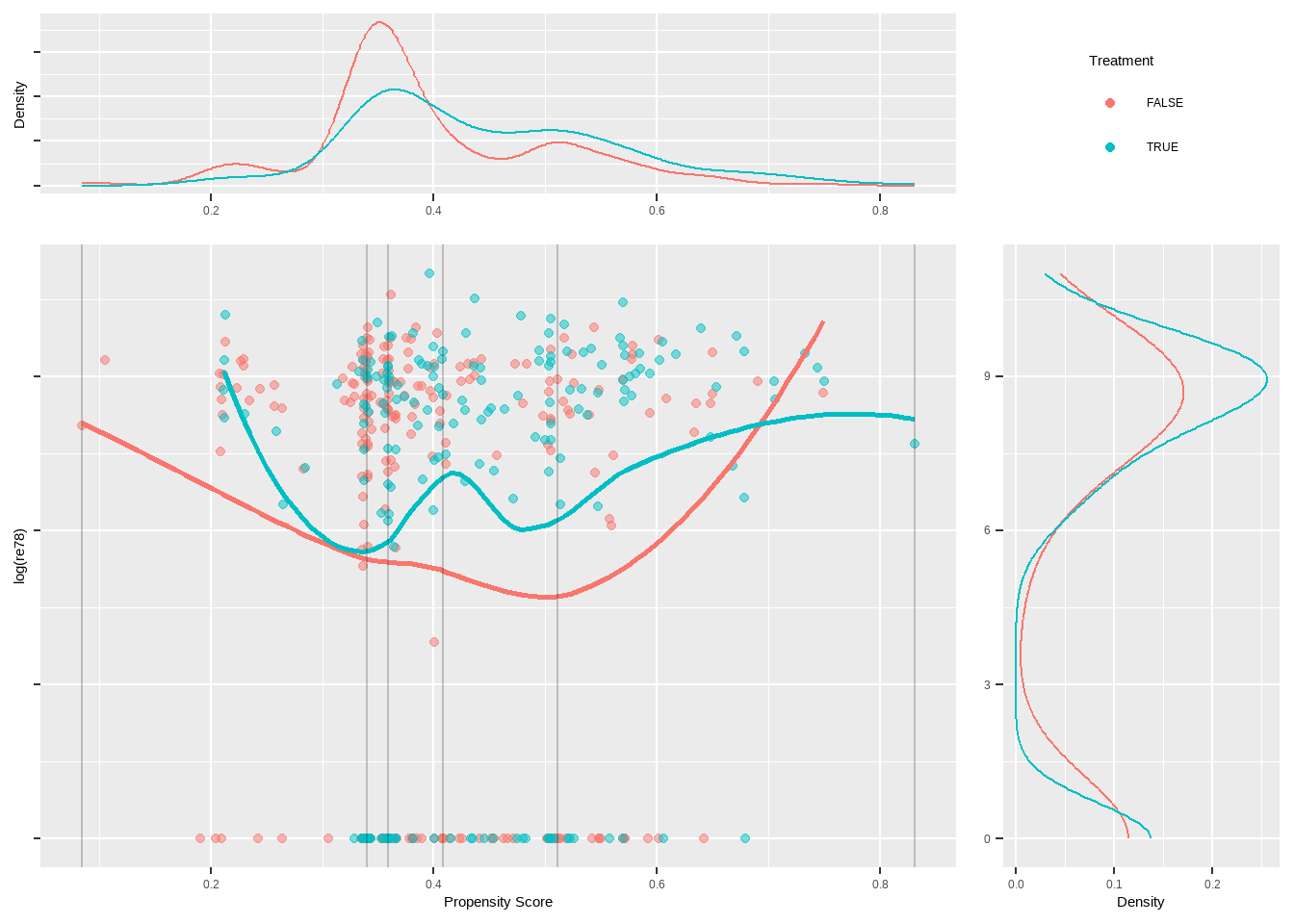
Show the code lalonde %>% ggplot ( aes ( x = ps , y = re78 , color = treat ) ) + geom_point ( alpha = 0.5 ) +
geom_smooth ( method = 'loess' , formula = y ~ x , alpha = 0.5 ) +
xlab ( 'Propensity Score' ) +
theme ( legend.position = 'bottom' )
具有大约 95% 置信区间(灰色)的 Loess 回归线在整个倾向得分范围内重叠,说明异质或不均匀
分层
Show the code lalonde $ treat <- as.numeric ( lalonde $ treat ) - 1 dat <- lalonde |> mutate ( ate_weight = psa :: calculate_ps_weights ( treat , ps , estimand = 'ATE' ) ,
att_weight = psa :: calculate_ps_weights ( treat , ps , estimand = 'ATT' ) ,
atc_weight = psa :: calculate_ps_weights ( treat , ps , estimand = 'ATC' ) ,
atm_weight = psa :: calculate_ps_weights ( treat , ps , estimand = 'ATM' )
) ggplot ( dat , aes ( x = ps , color = factor ( treat ) ) ) + geom_density ( aes ( fill = factor ( treat ) ) , alpha = 0.2 ) +
geom_vline ( xintercept = breaks5 $ breaks , alpha = 0.5 ) +
geom_text ( data = breaks5 $ labels ,
aes ( x = xmid , y = 0 , label = strata ) ,
color = 'black' , vjust = 0 , size = 4 ) +
xlab ( 'Propensity Score' ) + ylab ( 'Density' ) +
xlim ( c ( 0 , 1 ) )
Show the code PSAgraphics :: loess.psa ( response = log ( lalonde $ re78 + 1 ) , treatment = lalonde $ treat ,
propensity = lalonde $ ps ) #> $ATE
#> [1] 0.9008386
#>
#> $se.wtd
#> [1] 0.3913399
#>
#> $CI95
#> [1] 0.1181588 1.6835185
#>
#> $summary.strata
#> counts.0 counts.1 means.0 means.1 diff.means
#> 1 34 11 6.268705 6.474912 0.2062076
#> 2 32 12 5.491717 5.659280 0.1675631
#> 3 31 14 5.467712 5.703584 0.2358722
#> 4 30 14 5.425593 5.747613 0.3220194
#> 5 27 18 5.397146 5.831117 0.4339703
#> 6 24 20 5.302660 6.339721 1.0370619
#> 7 21 23 5.125331 6.607936 1.4826043
#> 8 21 24 5.036908 6.594808 1.5578999
#> 9 22 22 5.182703 6.981383 1.7986801
#> 10 18 27 6.047529 7.820786 1.7732573
psa::loess_plot(ps = lalonde$ps,
outcome = log(lalonde$re78 + 1),
treatment = lalonde$treat == 1,
outcomeTitle = 'log(re78)',
plot.strata = 5,
points.treat.alpha = 0.5,
points.control.alpha = 0.5,
percentPoints.treat = 1,
percentPoints.control = 1,
se = FALSE,
method = 'loess')
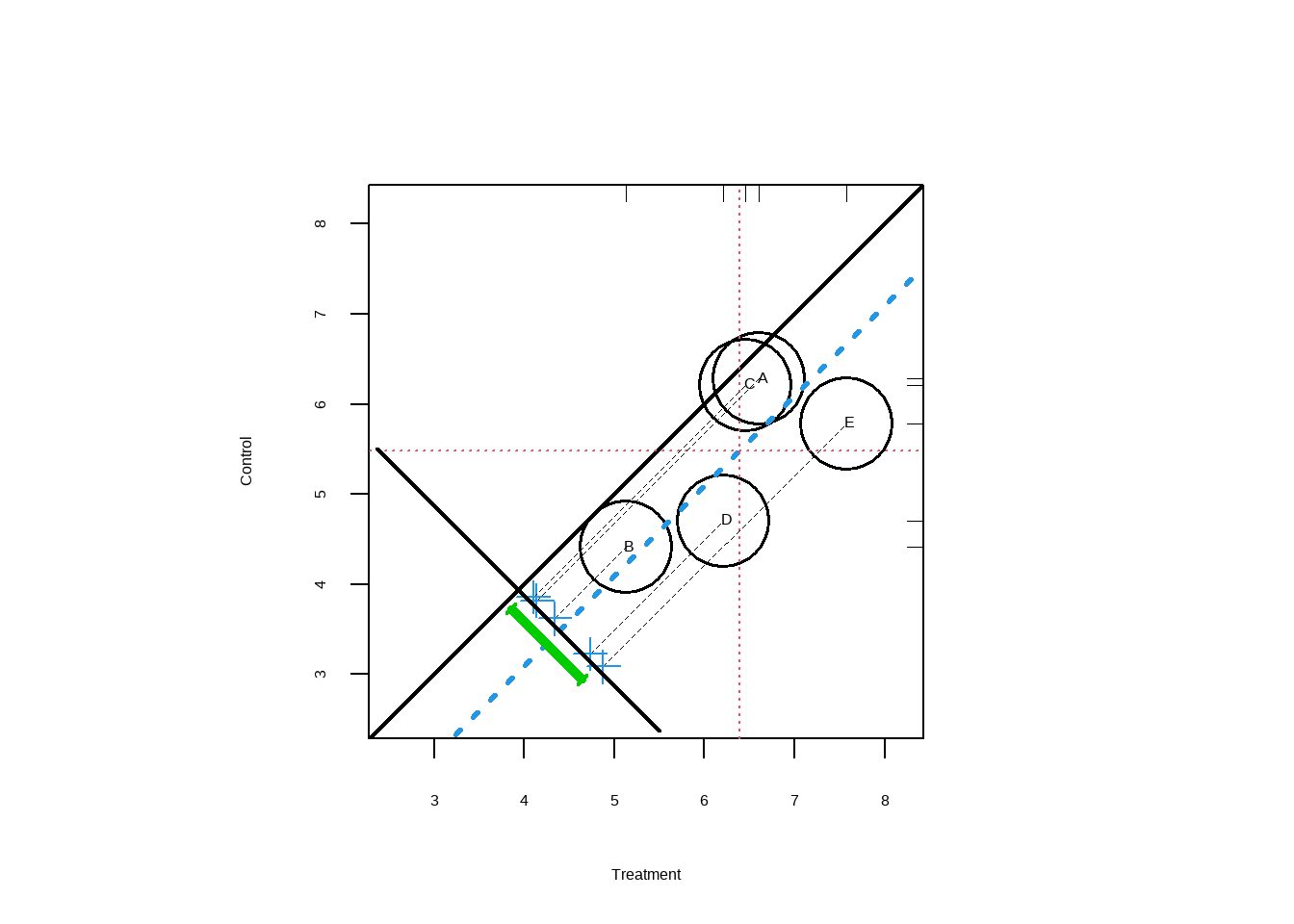
Show the code PSAgraphics :: circ.psa ( response = log ( lalonde $ re78 + 1 ) , treatment = lalonde $ treat == 1 ,
strata = lalonde $ lr_strata5 ,
revc = T ,
xlab = 'Treatment' ,
ylab = 'Control' ) #> $summary.strata
#> n.FALSE n.TRUE means.FALSE means.TRUE
#> A 66 23 6.280406 6.600537
#> B 61 28 4.409935 5.129193
#> C 51 38 6.212981 6.455034
#> D 42 47 4.705981 6.208840
#> E 40 49 5.783529 7.576461
#>
#> $wtd.Mn.TRUE
#> [1] 6.394013
#>
#> $wtd.Mn.FALSE
#> [1] 5.478567
#>
#> $ATE
#> [1] -0.9154463
#>
#> $se.wtd
#> [1] 0.394155
#>
#> $approx.t
#> [1] -2.322554
#>
#> $df
#> [1] 435
#>
#> $CI.95
#> [1] -1.6901314 -0.1407612
将绘制每个层的平均处理(x 轴)与对照(y 轴)的对比。均值被投影到垂直于单位线的线(即线y=x), 以便刻度线表示差异的分布。绿色条对应于 95% 置信区间。圆圈的大小与每个层内的样本大小成正比。 因为y=x落在该线上的点表示零差(即y−x=0). 推而广之,如果绿线表示的置信区间跨越单位线,则无法否定原假设。然而,在此示例中,存在统计学上显著的处理效果。
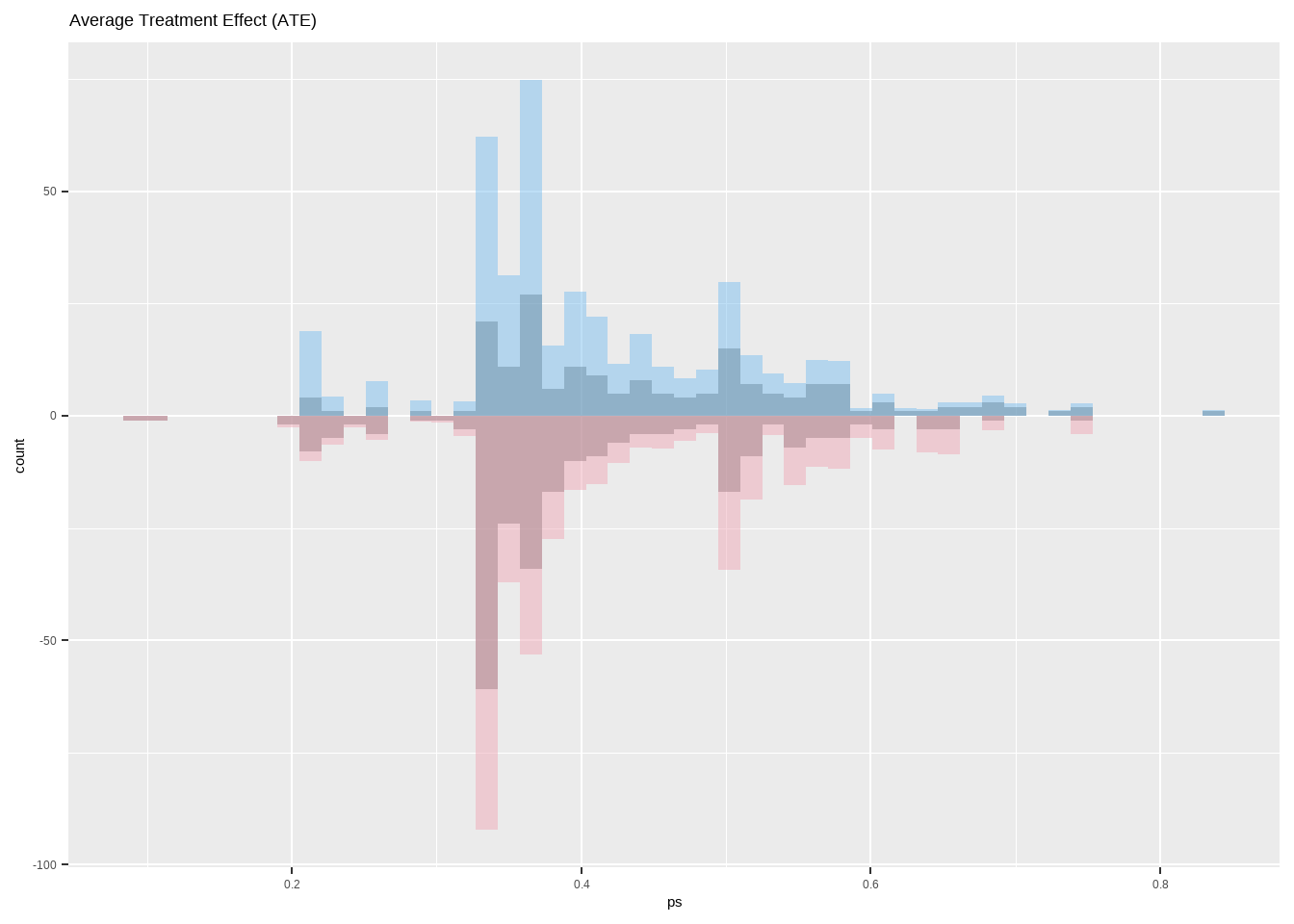
平均治疗效应ATE
Show the code ggplot ( ) + geom_histogram ( data = dat [ dat $ treat == 1 ,] ,
aes ( x = ps , y = after_stat ( count ) ) ,
bins = 50 , alpha = 0.5 ) +
geom_histogram ( data = dat [ dat $ treat == 1 ,] ,
aes ( x = ps , weight = ate_weight , y = after_stat ( count ) ) ,
bins = 50 ,
fill = "skyblue2" , alpha = 0.5 ) +
geom_histogram ( data = dat [ dat $ treat == 0 ,] ,
aes ( x = ps , y = - after_stat ( count ) ) ,
bins = 50 , alpha = 0.5 ) +
geom_histogram ( data = dat [ dat $ treat == 0 ,] ,
aes ( x = ps , weight = ate_weight , y = - after_stat ( count ) ) ,
bins = 50 ,
fill = "pink2" , alpha = 0.5 ) +
ggtitle ( 'Average Treatment Effect (ATE)' )
接受治疗者的平均治疗效果 (ATT)
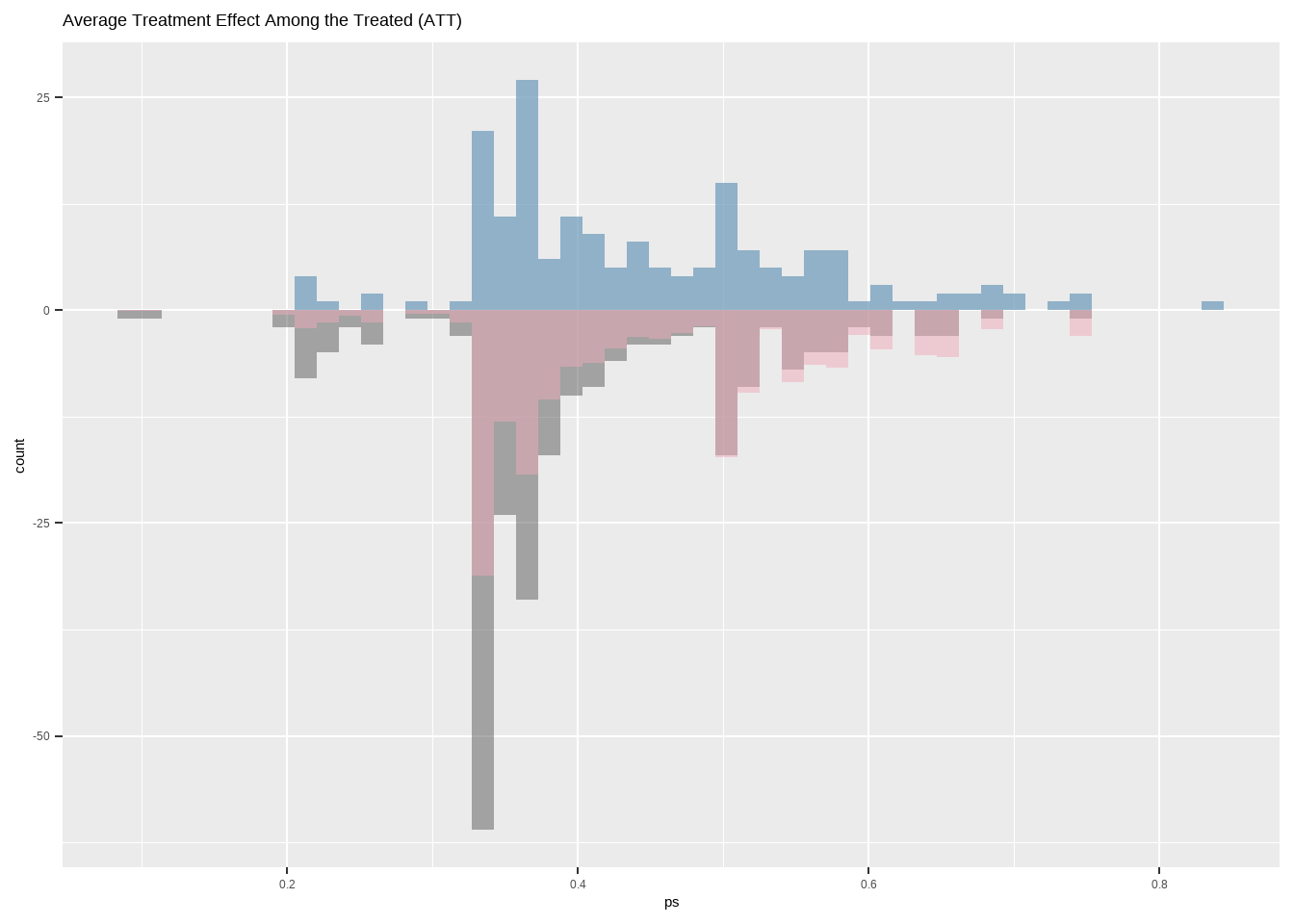
Show the code ggplot ( ) + geom_histogram ( data = dat [ dat $ treat == 1 ,] ,
aes ( x = ps , y = after_stat ( count ) ) ,
bins = 50 , alpha = 0.5 ) +
geom_histogram ( data = dat [ dat $ treat == 1 ,] ,
aes ( x = ps , weight = att_weight , y = after_stat ( count ) ) ,
bins = 50 ,
fill = "skyblue2" , alpha = 0.5 ) +
geom_histogram ( data = dat [ dat $ treat == 0 ,] ,
aes ( x = ps , y = - after_stat ( count ) ) ,
bins = 50 , alpha = 0.5 ) +
geom_histogram ( data = dat [ dat $ treat == 0 ,] ,
aes ( x = ps , weight = att_weight , y = - after_stat ( count ) ) ,
bins = 50 ,
fill = "pink2" , alpha = 0.5 ) +
ggtitle ( 'Average Treatment Effect Among the Treated (ATT)' )
对照组的平均治疗效果 (ATC)
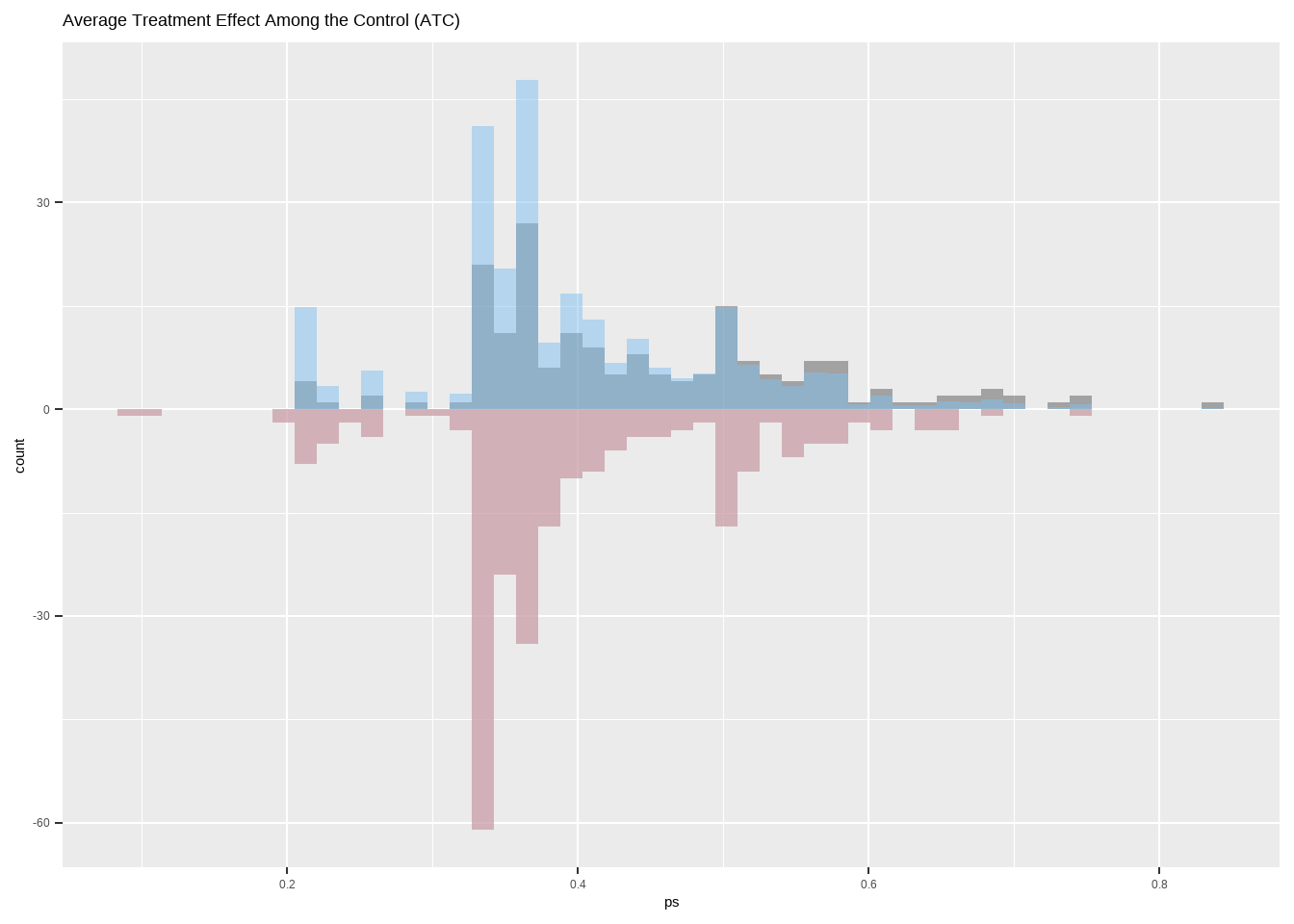
Show the code ggplot ( ) + geom_histogram ( data = dat [ dat $ treat == 1 ,] ,
aes ( x = ps , y = after_stat ( count ) ) ,
bins = 50 , alpha = 0.5 ) +
geom_histogram ( data = dat [ dat $ treat == 1 ,] ,
aes ( x = ps , weight = atc_weight , y = after_stat ( count ) ) ,
bins = 50 ,
fill = "skyblue2" , alpha = 0.5 ) +
geom_histogram ( data = dat [ dat $ treat == 0 ,] ,
aes ( x = ps , y = - after_stat ( count ) ) ,
bins = 50 , alpha = 0.5 ) +
geom_histogram ( data = dat [ dat $ treat == 0 ,] ,
aes ( x = ps , weight = atc_weight , y = - after_stat ( count ) ) ,
bins = 50 ,
fill = "pink" , alpha = 0.5 ) +
ggtitle ( 'Average Treatment Effect Among the Control (ATC)' )
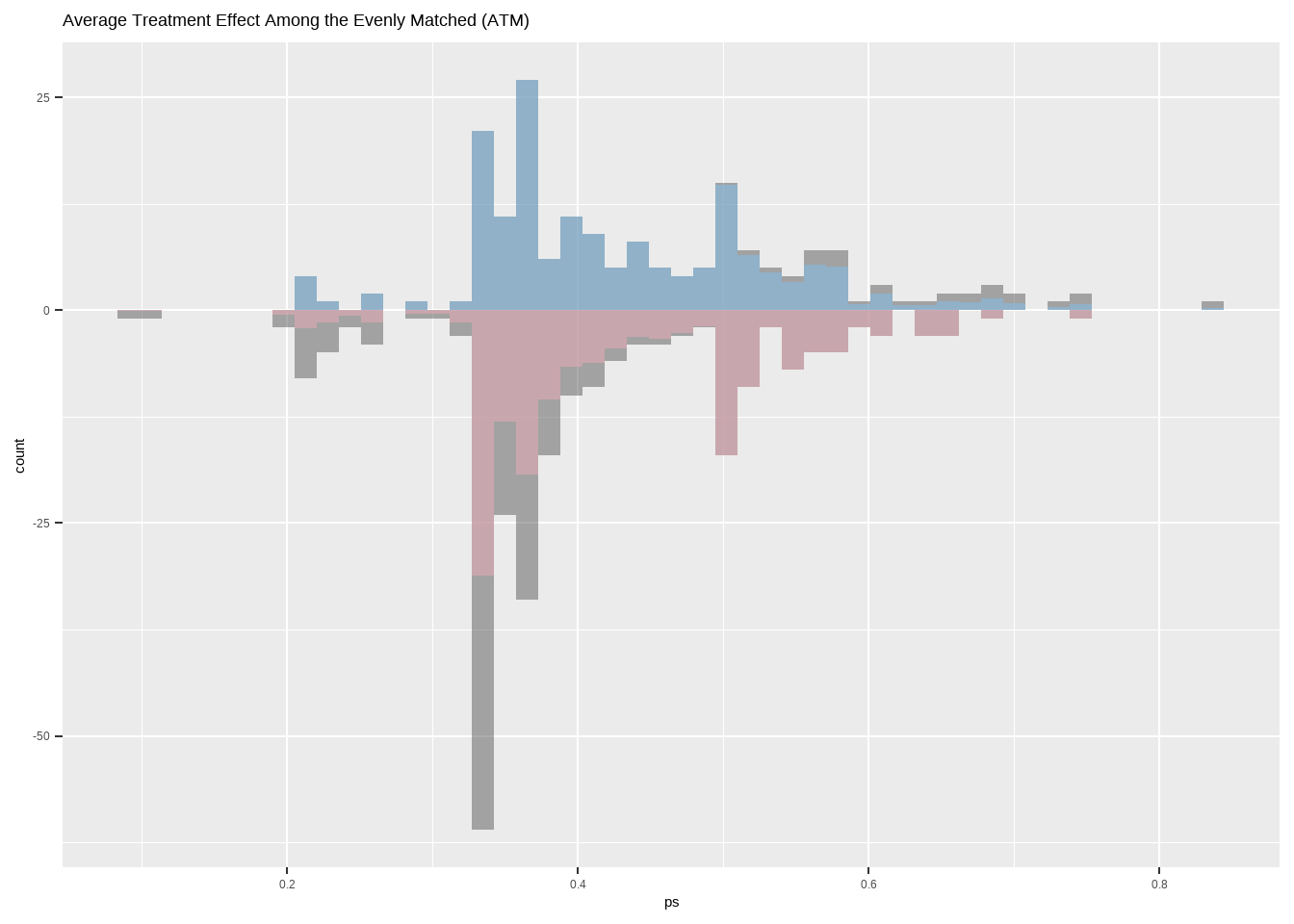
Average Treatment Effect Among the Evenly Matched (ATM)
Show the code ggplot ( ) + geom_histogram ( data = dat [ dat $ treat == 1 ,] ,
aes ( x = ps , y = after_stat ( count ) ) ,
bins = 50 , alpha = 0.5 ) +
geom_histogram ( data = dat [ dat $ treat == 1 ,] ,
aes ( x = ps , weight = atm_weight , y = after_stat ( count ) ) ,
bins = 50 ,
fill = "skyblue2" , alpha = 0.5 ) +
geom_histogram ( data = dat [ dat $ treat == 0 ,] ,
aes ( x = ps , y = - after_stat ( count ) ) ,
bins = 50 , alpha = 0.5 ) +
geom_histogram ( data = dat [ dat $ treat == 0 ,] ,
aes ( x = ps , weight = atm_weight , y = - after_stat ( count ) ) ,
bins = 50 ,
fill = "pink2" , alpha = 0.5 ) +
ggtitle ( 'Average Treatment Effect Among the Evenly Matched (ATM)' )
敏感性分析
评估该效果的稳健性
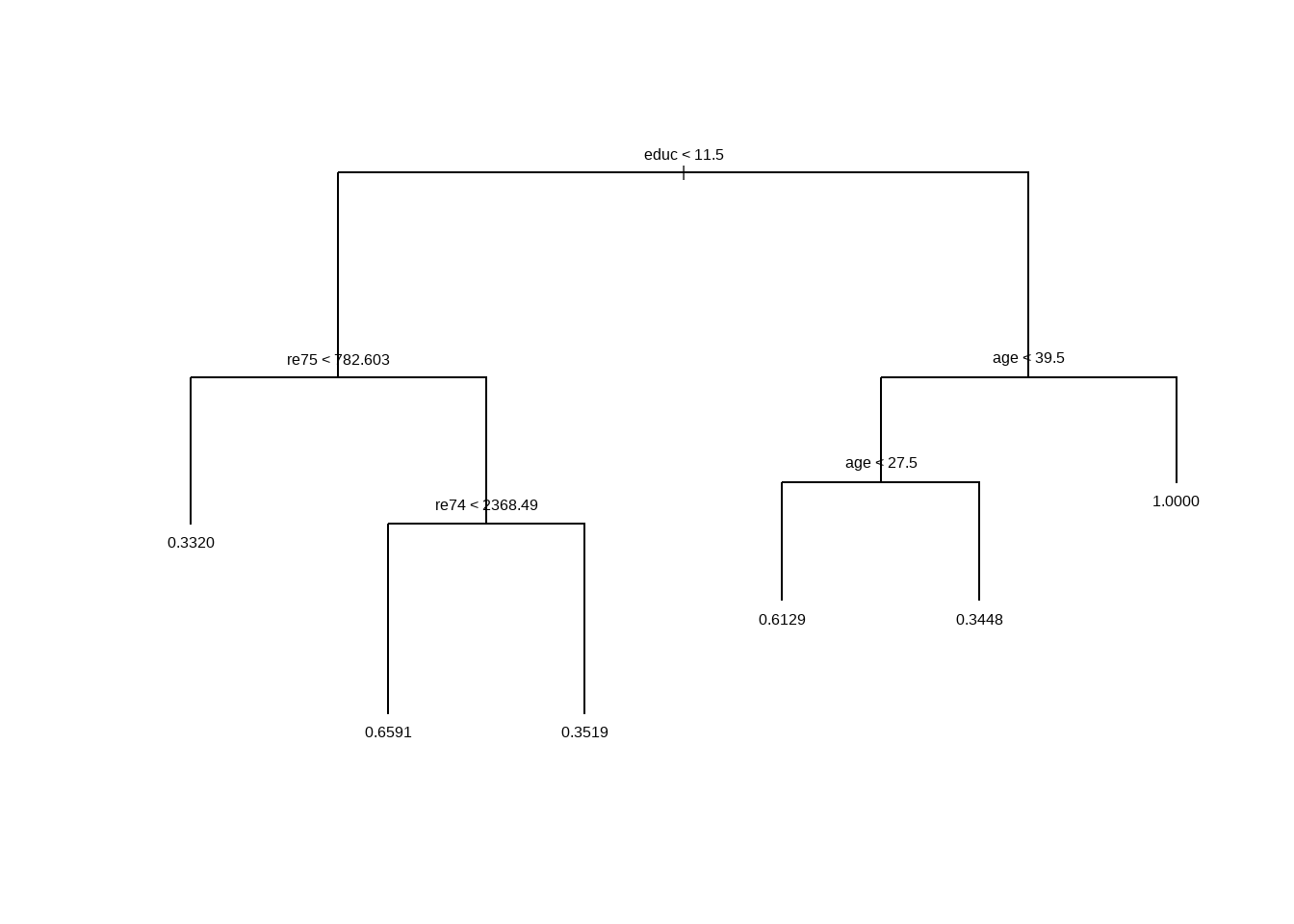
Rosenbaum (2012) 建议另一种测试灵敏度的方法是测试原假设两次。我们将在这里使用分类树方法来估计倾向得分和分层。
Show the code library ( tree ) tree_out <- tree :: tree ( lalonde_formula , data = lalonde )
plot ( tree_out ) ; text ( tree_out ) Show the code lalonde $ tree_ps <- predict ( tree_out ) table ( lalonde $ tree_ps , lalonde $ treat , useNA = 'ifany' ) #> #> 0 1 #> 0.332 167 83 #> 0.344827586206897 19 10 #> 0.351851851851852 35 19 #> 0.612903225806452 24 38 #> 0.659090909090909 15 29 #> 1 0 6 lalonde $ tree_strata <- predict ( tree_out , type = 'where' ) table ( lalonde $ tree_strata , lalonde $ treat , useNA = 'ifany' ) #> #> 0 1 #> 3 167 83 #> 5 15 29 #> 6 35 19 #> 9 24 38 #> 10 19 10 #> 11 0 6