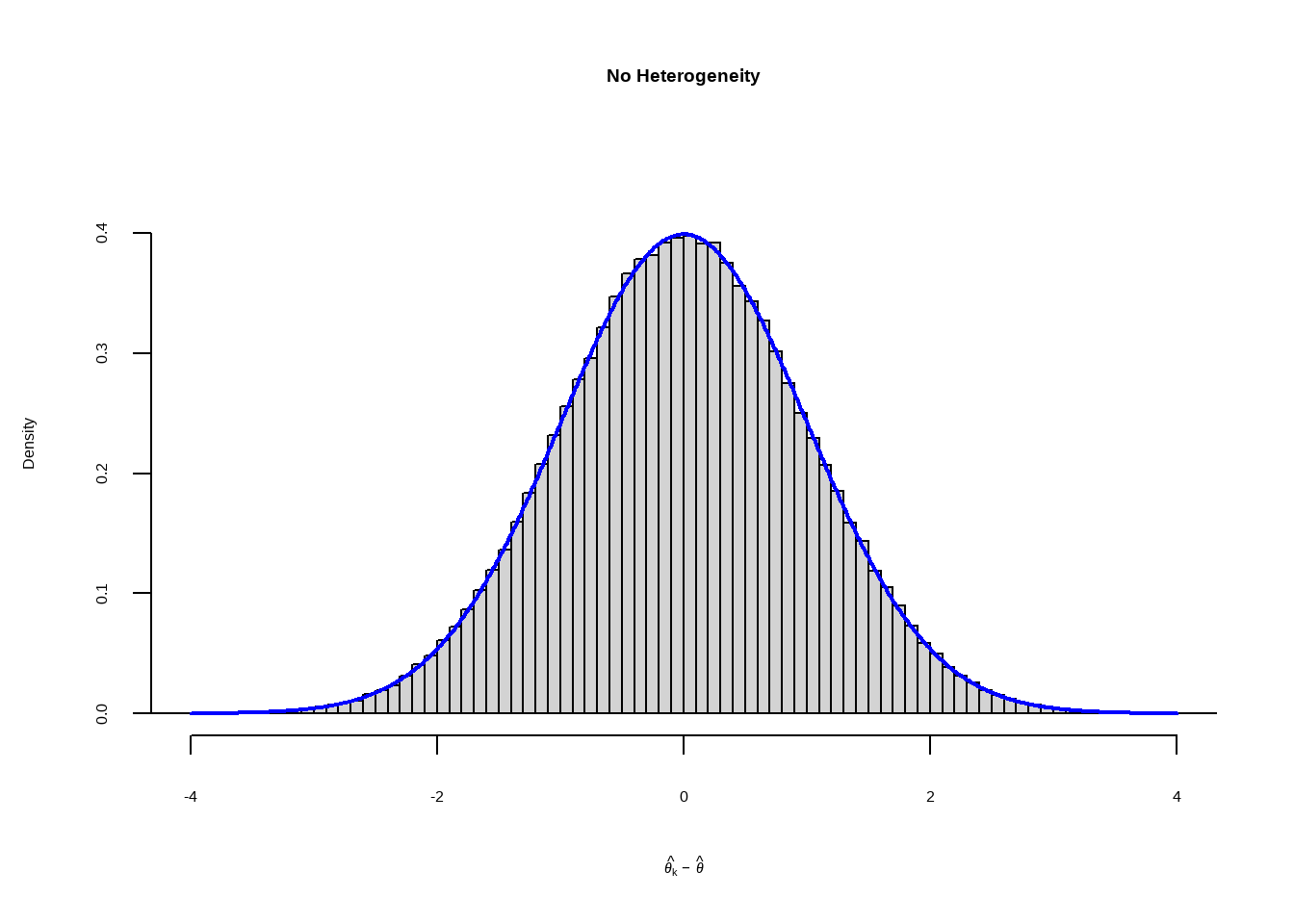
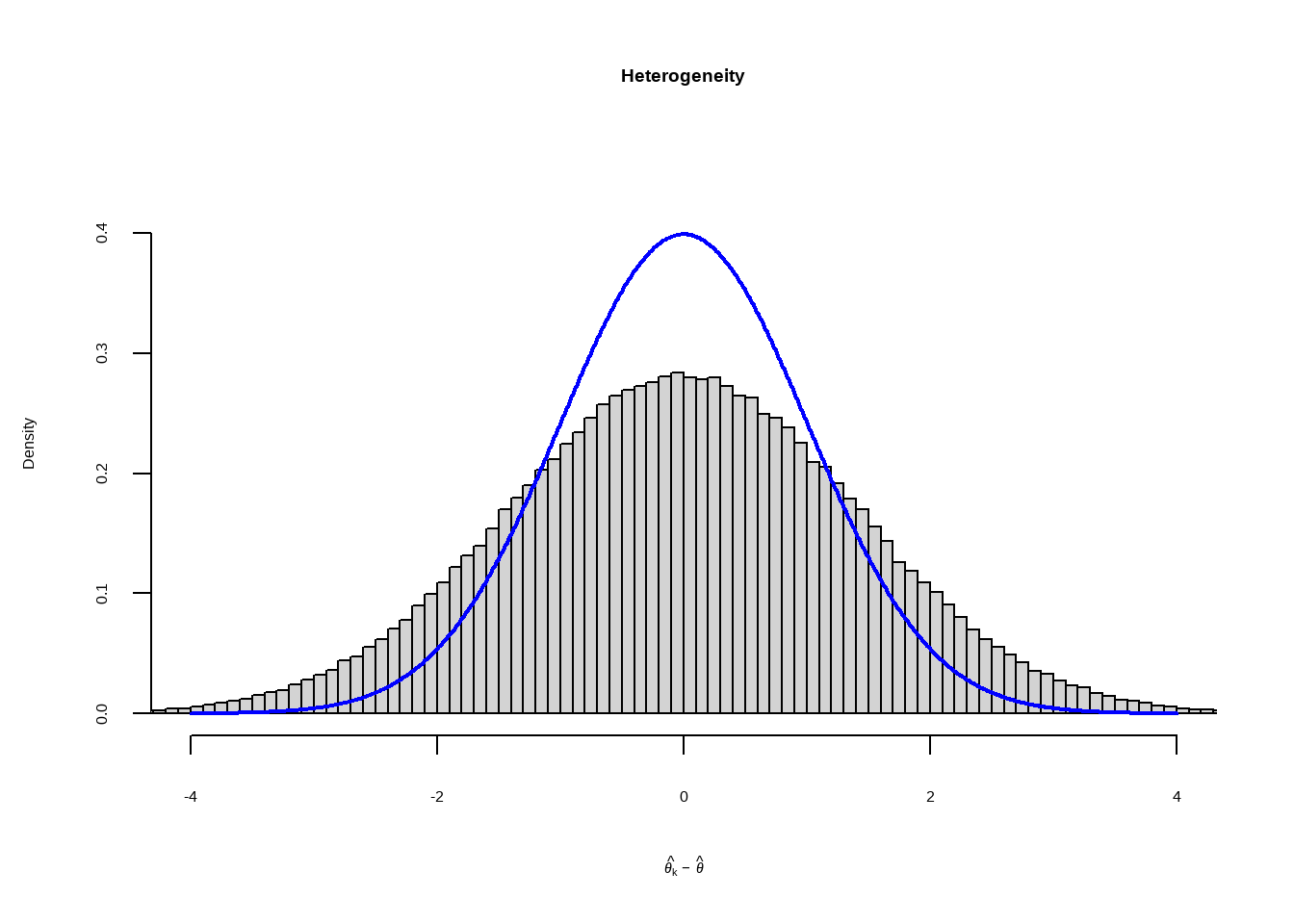
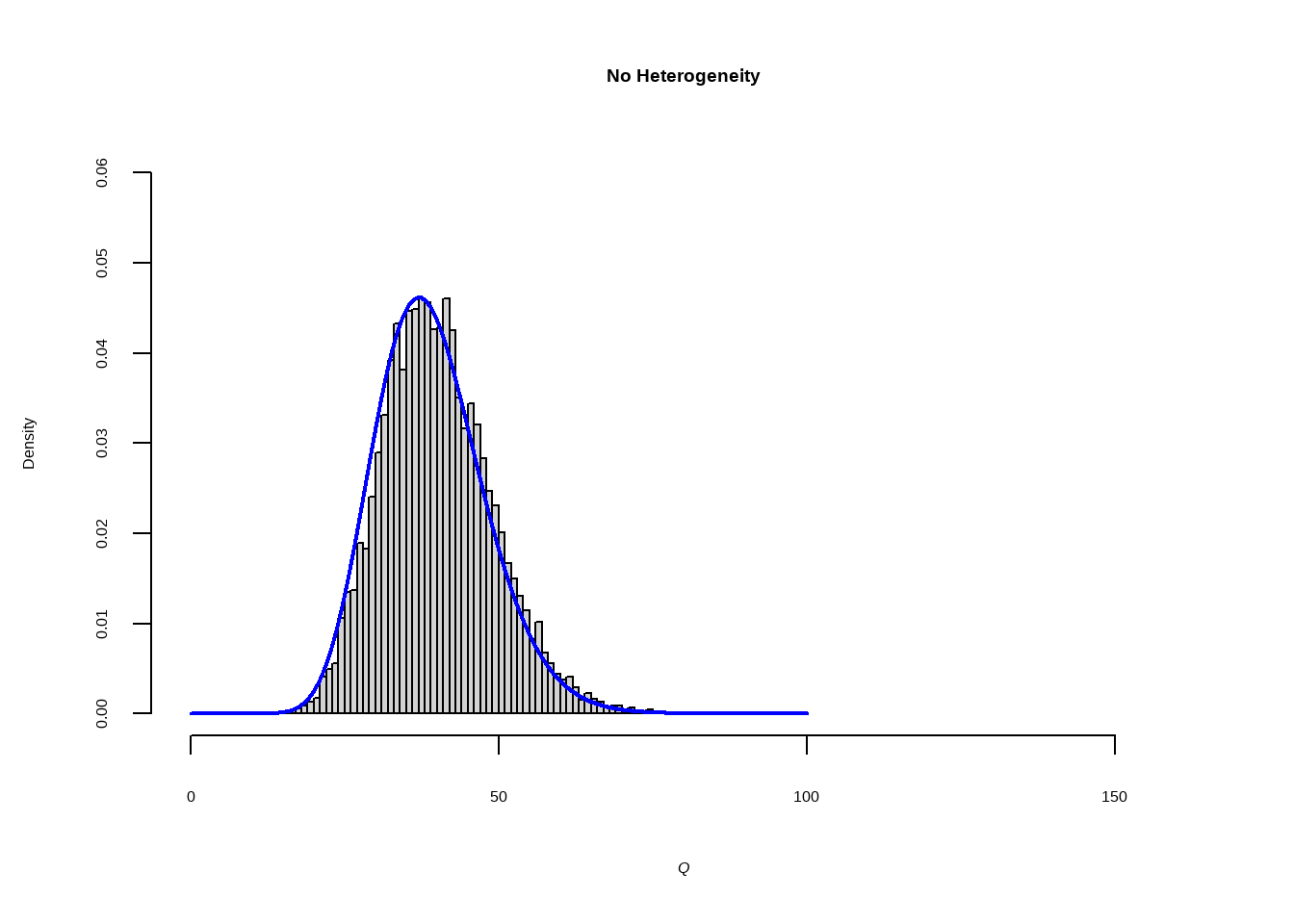
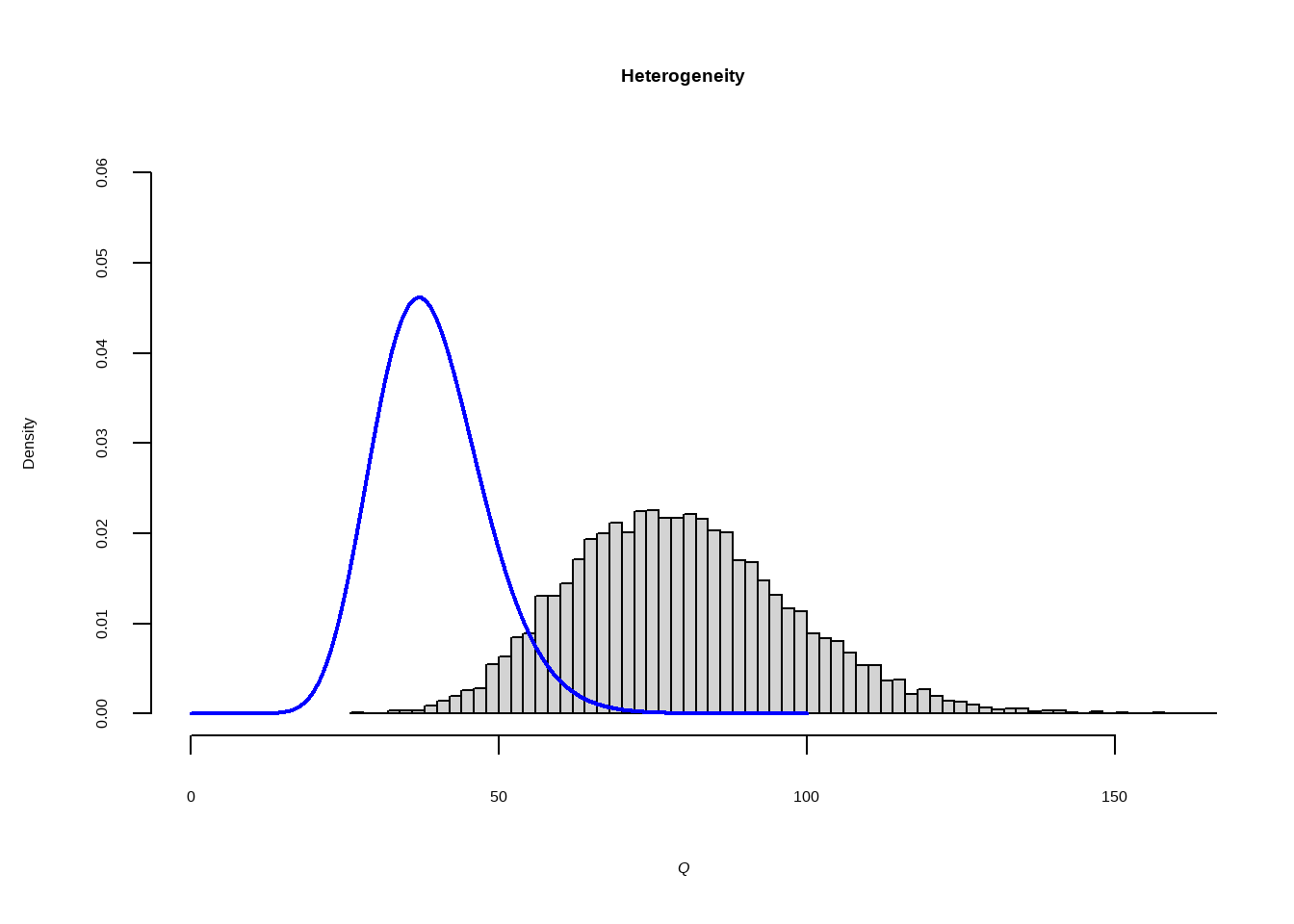
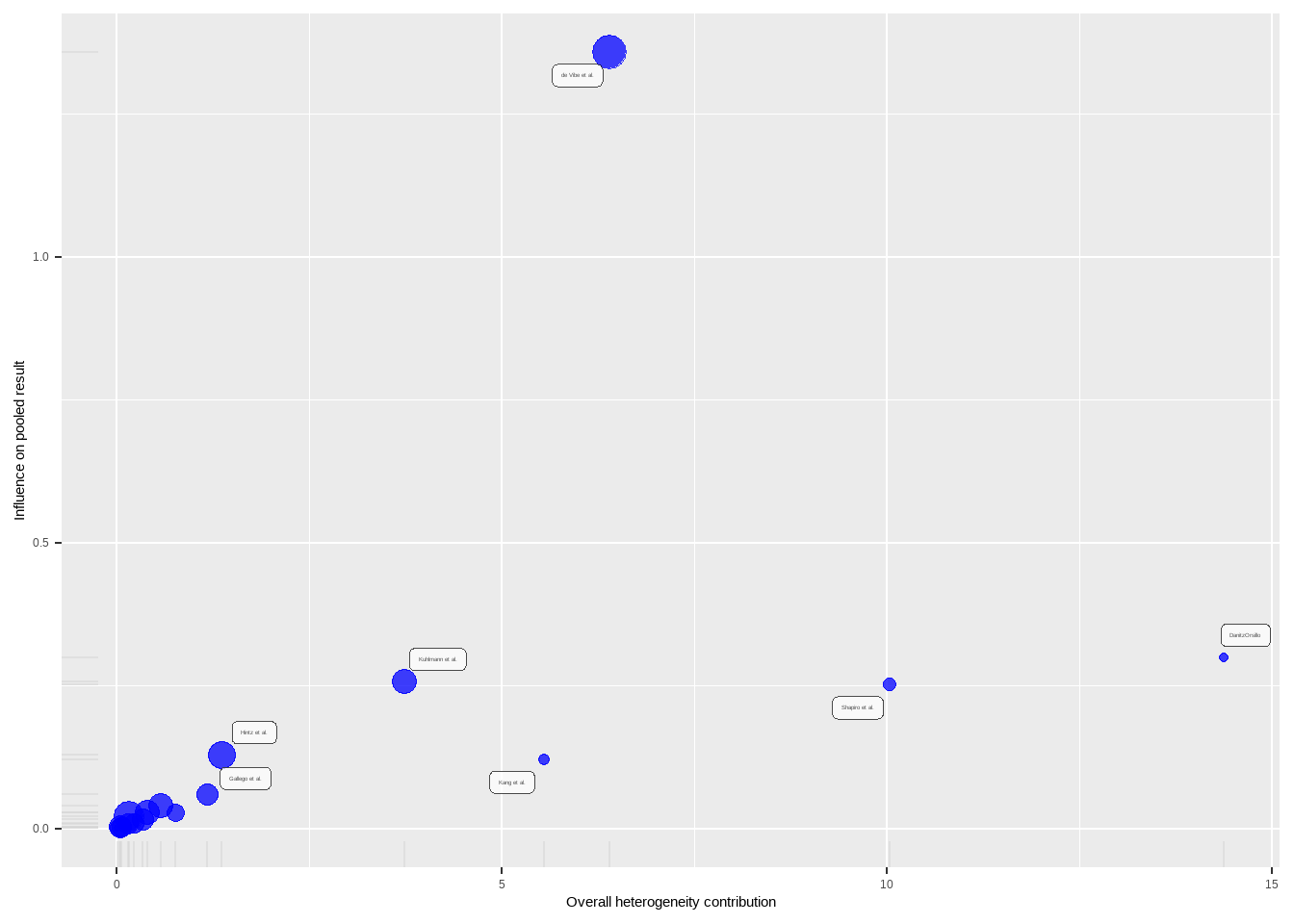
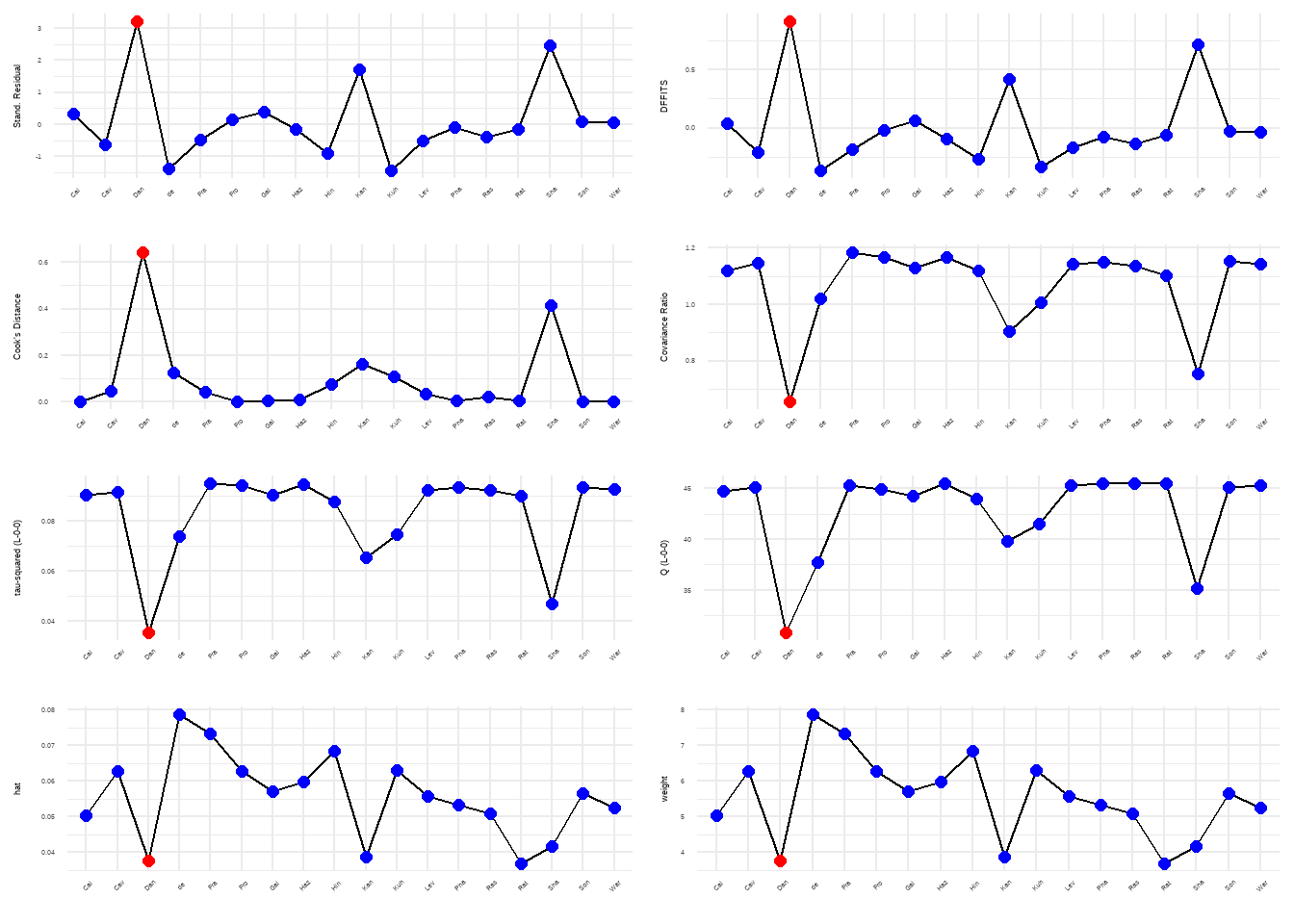
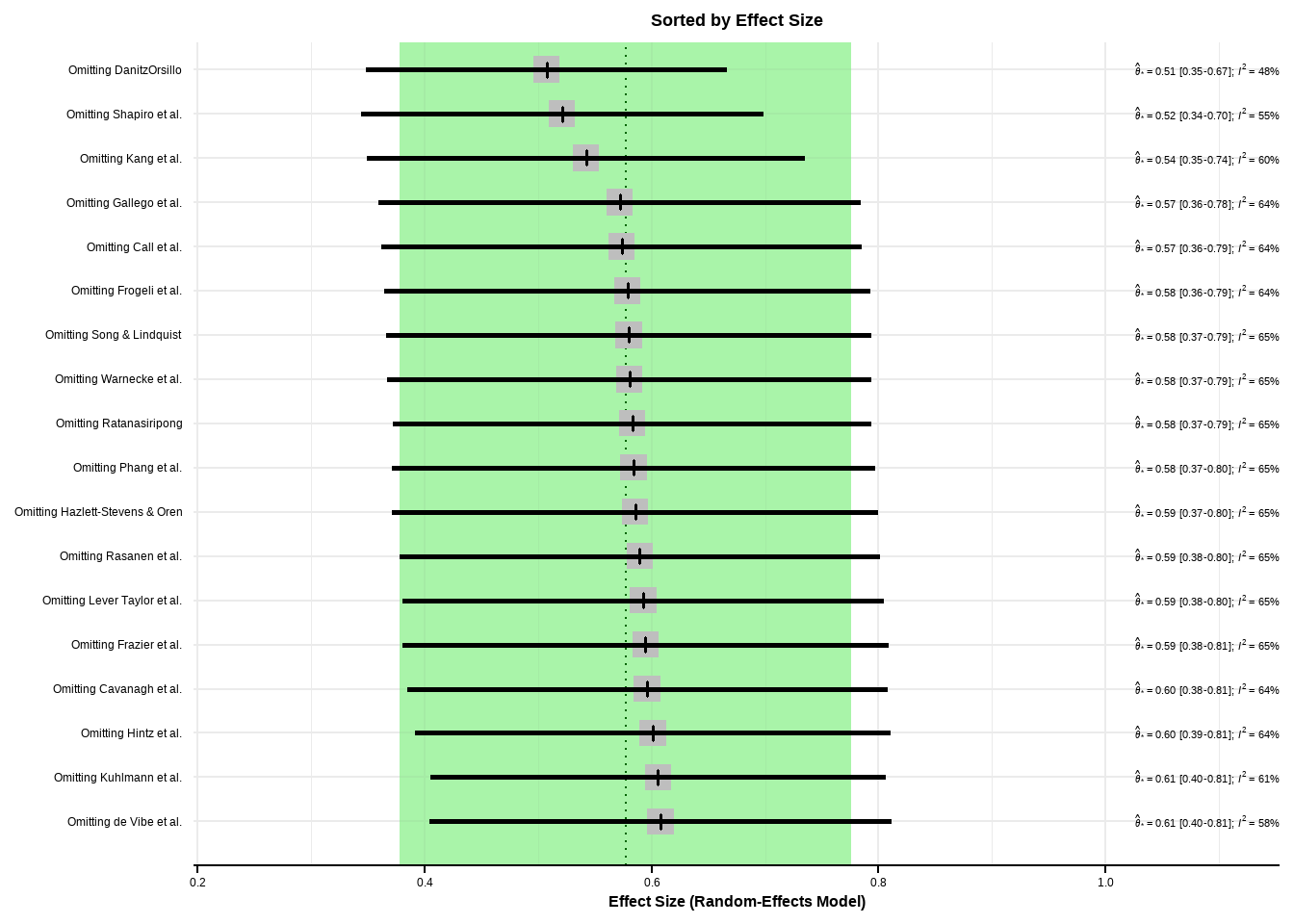
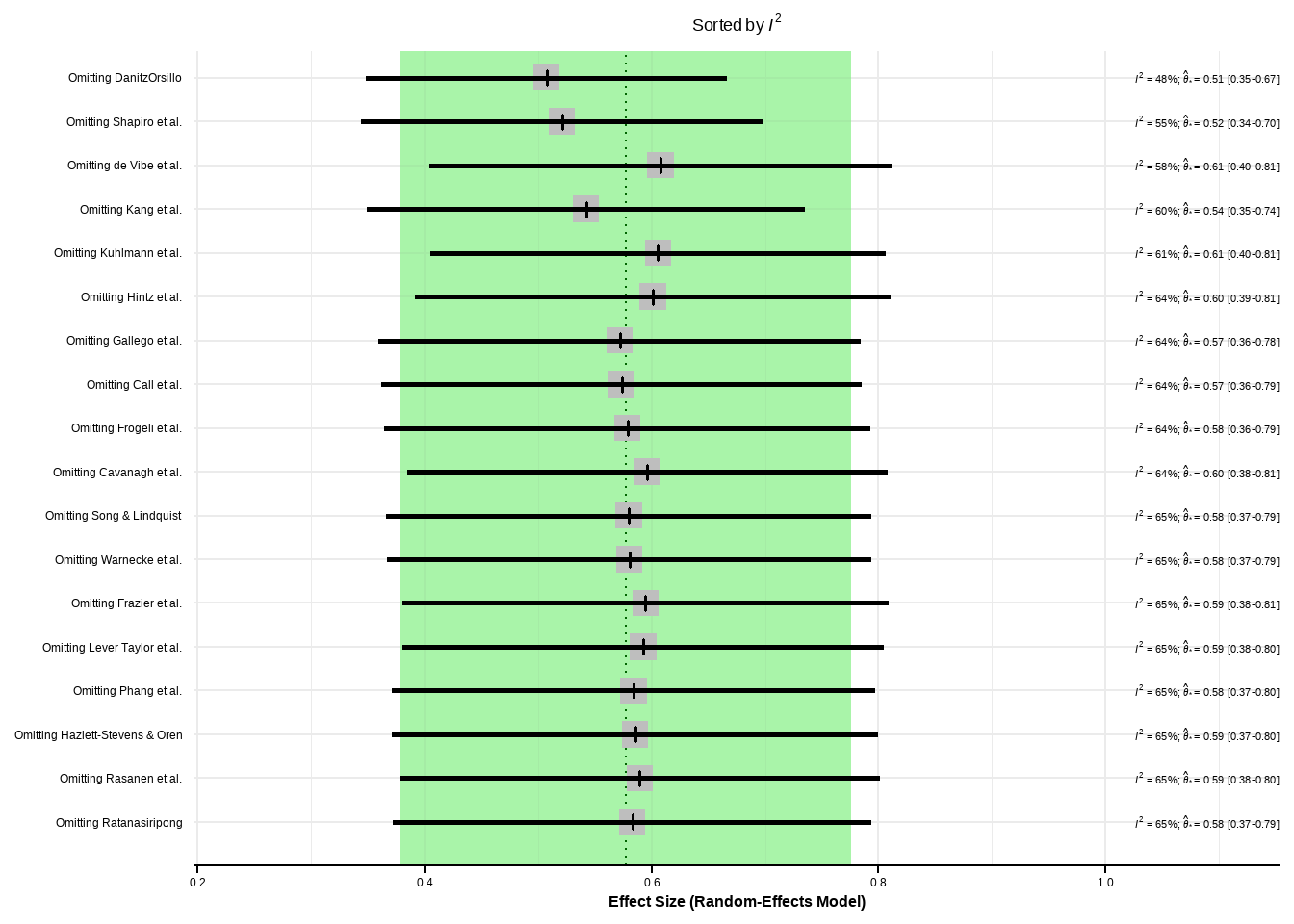
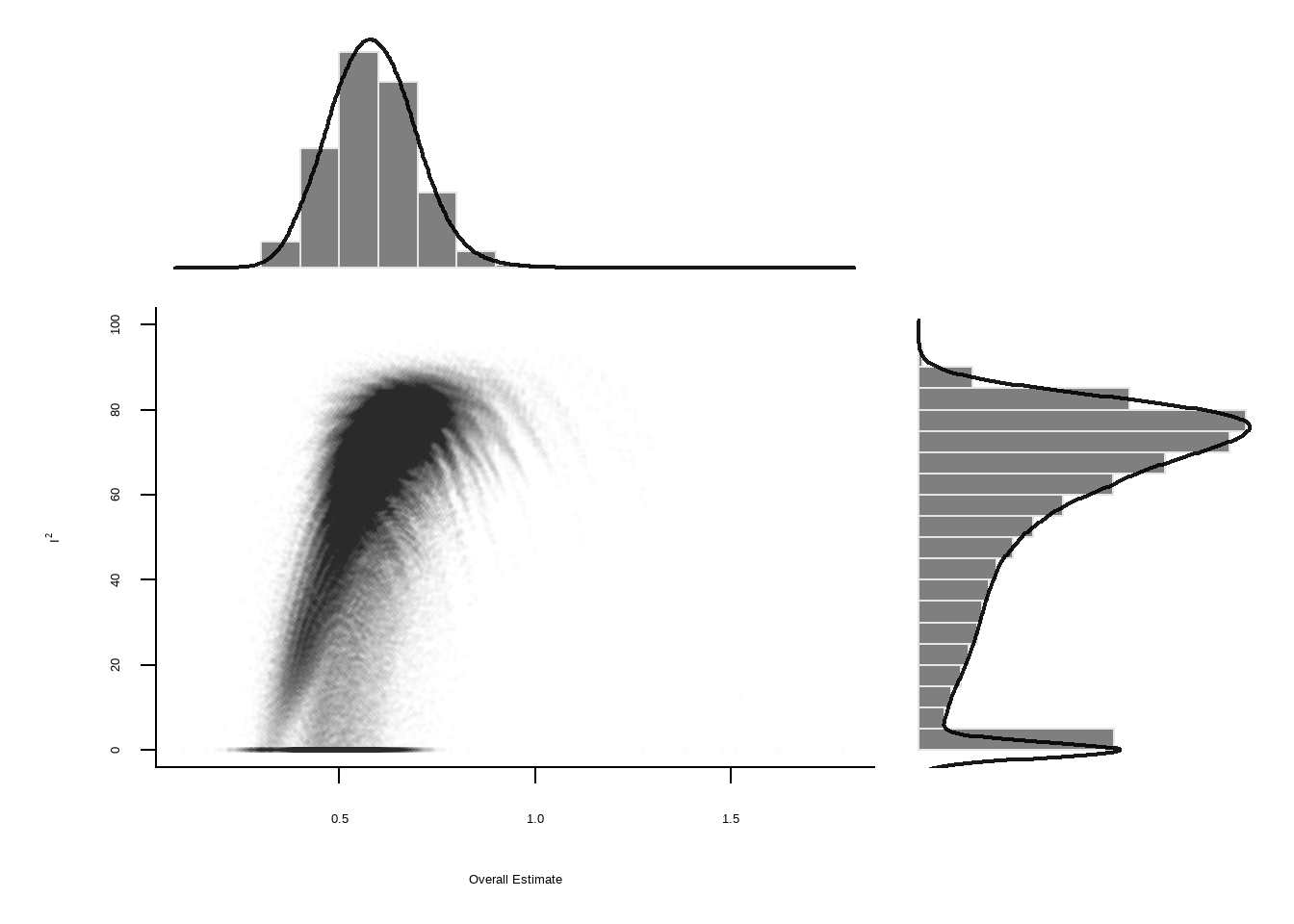
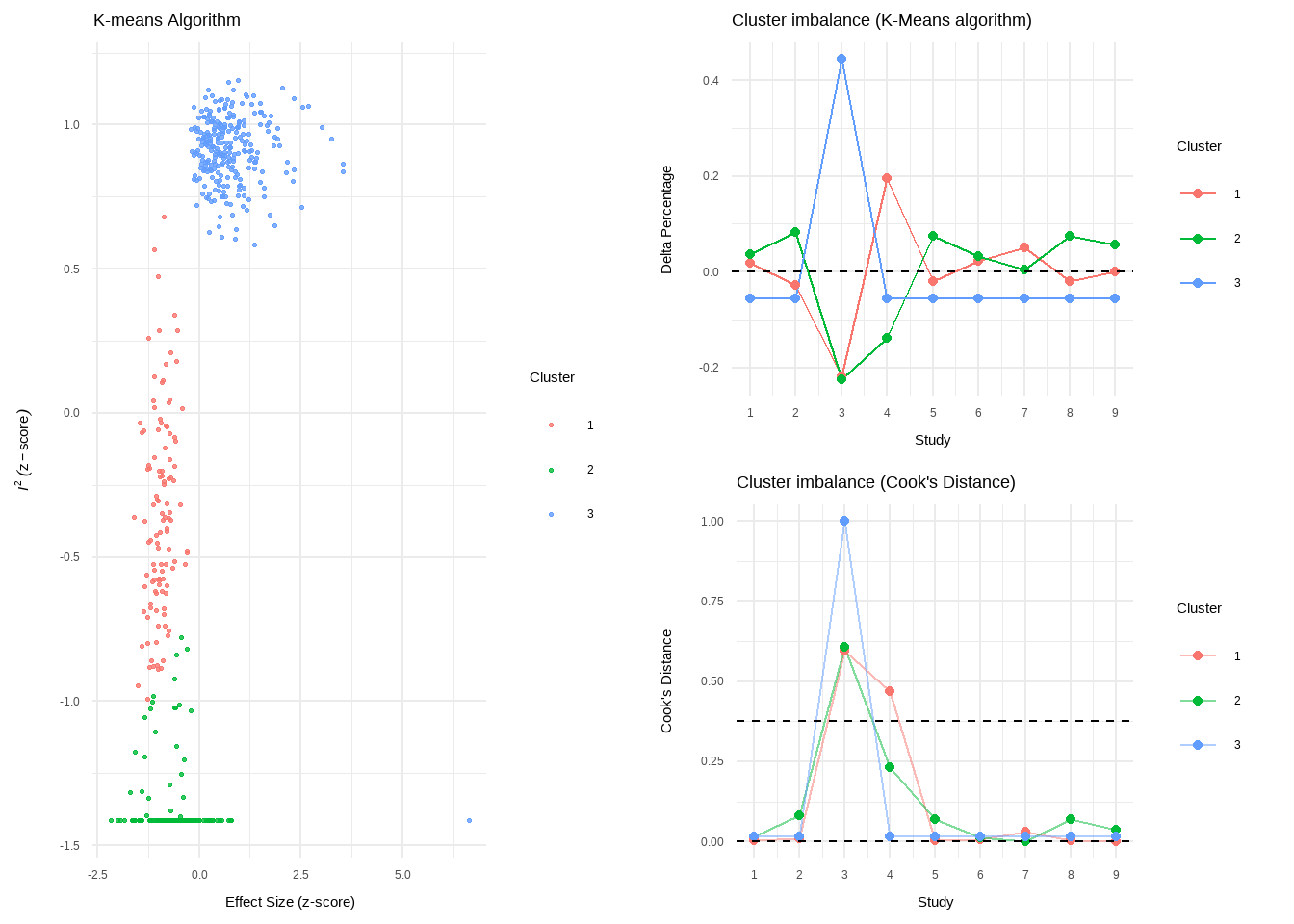
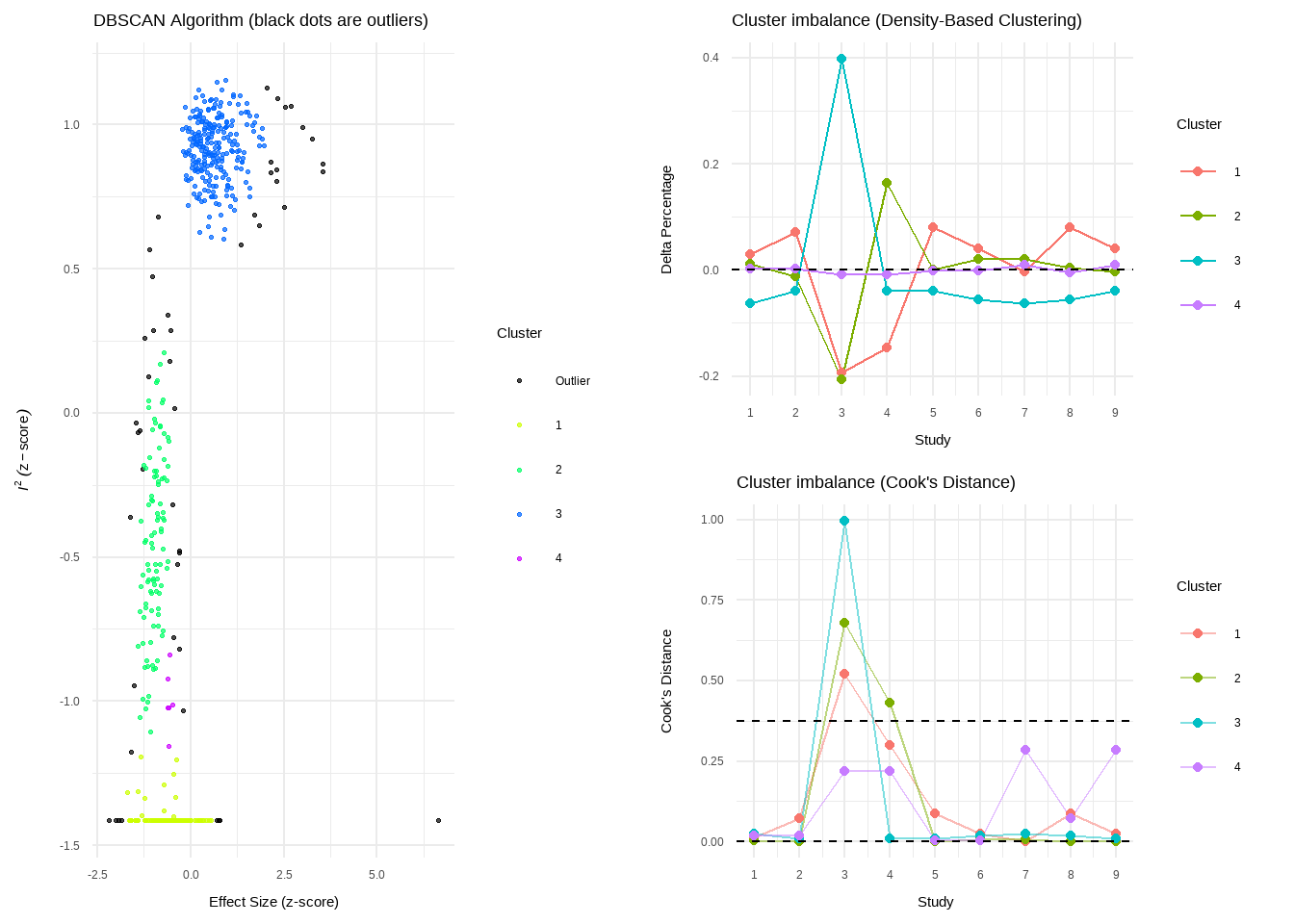
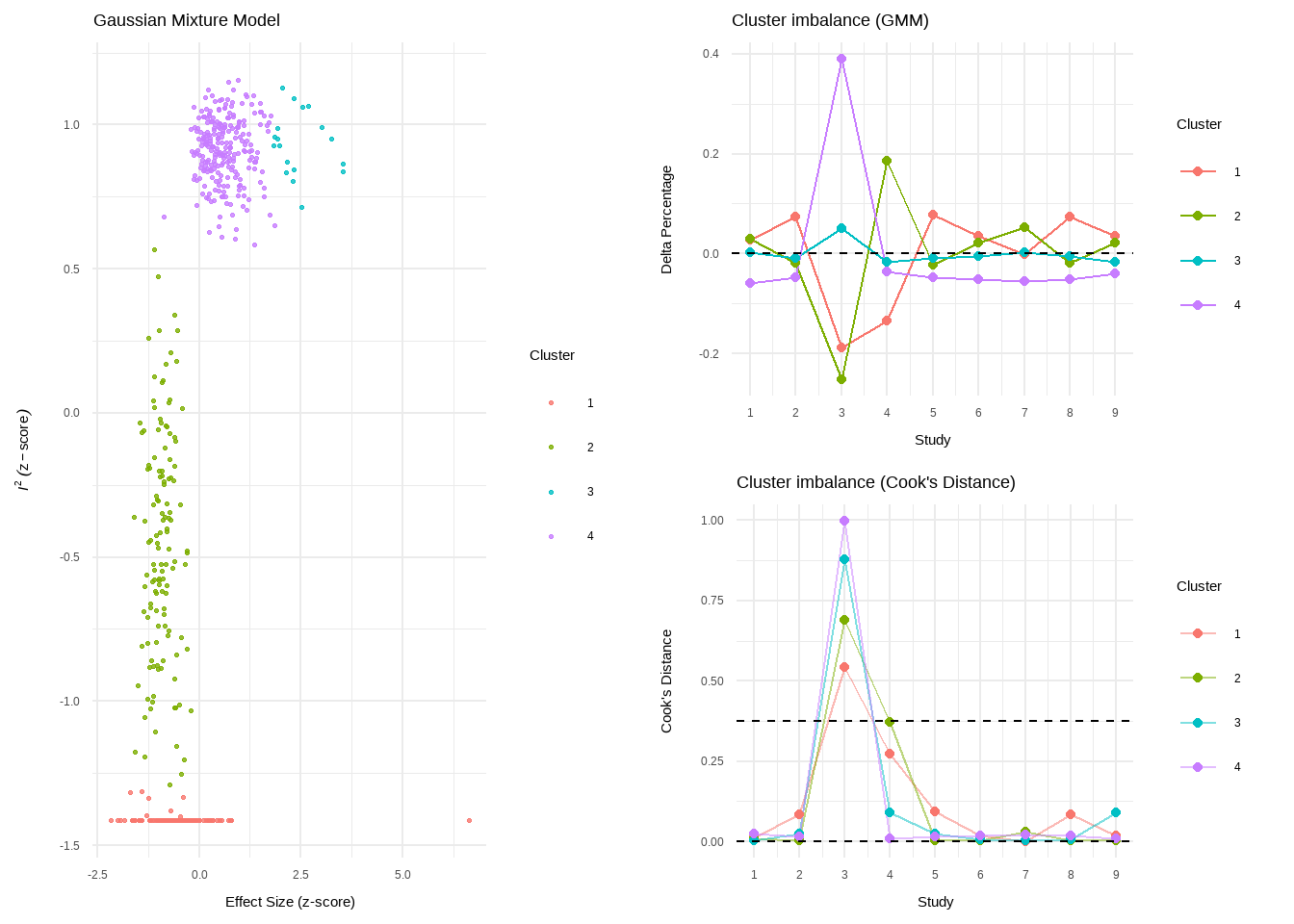
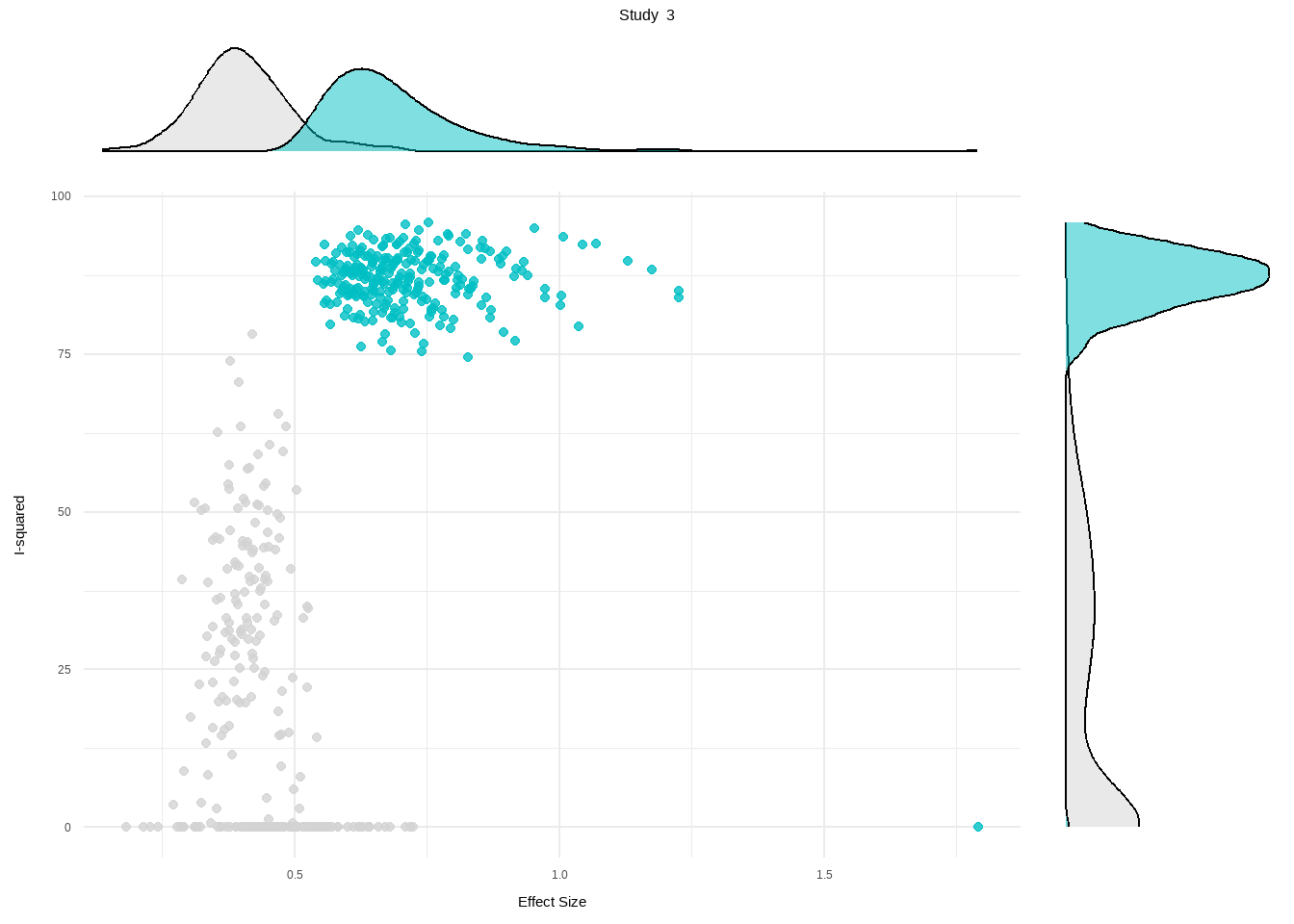
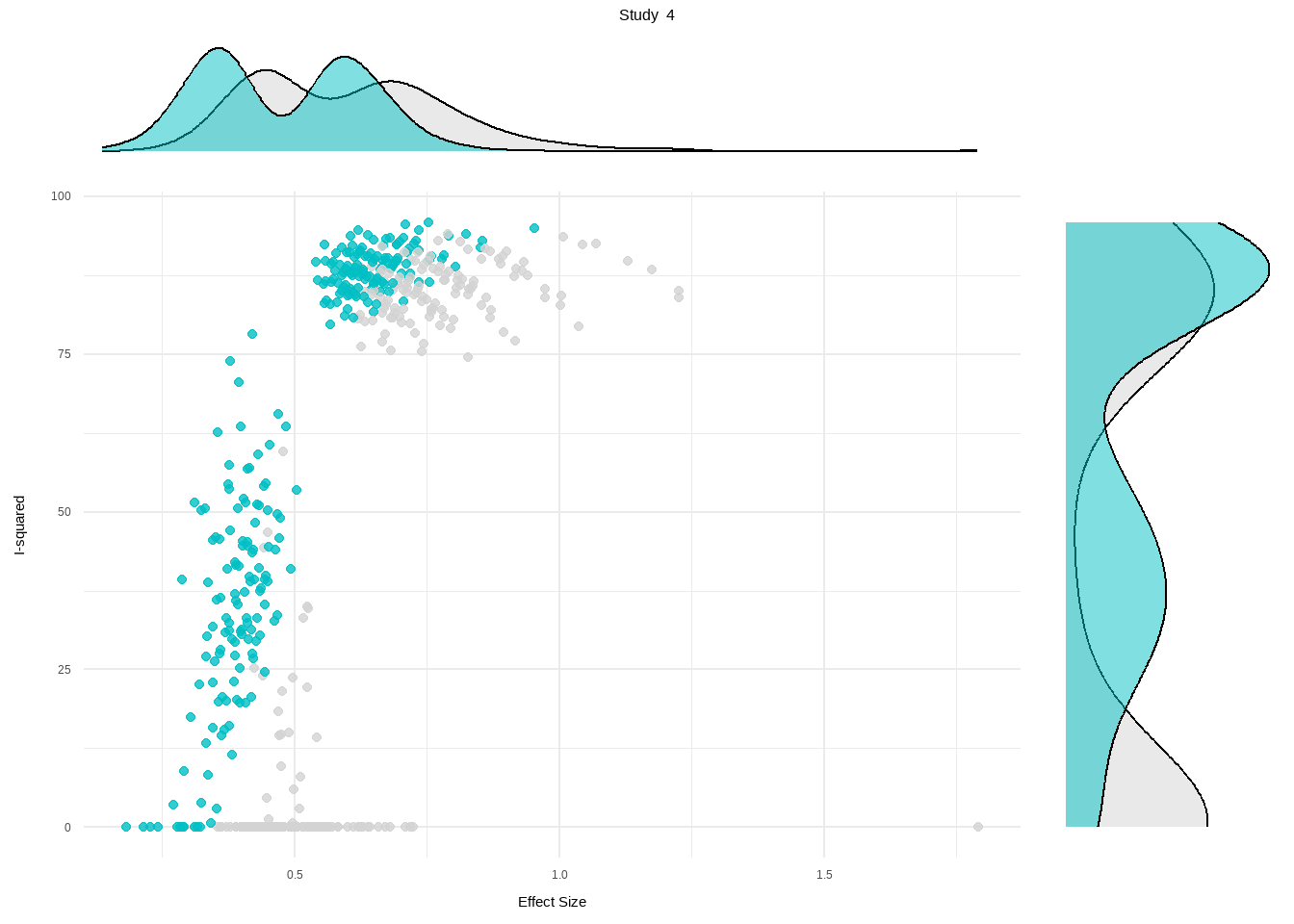
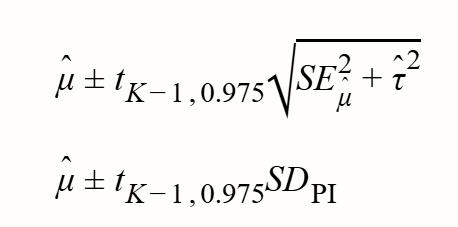--- title: "研究间异质性" --- ## Cochran's Q ```{r} set.seed (123 ) # needed to reproduce results rnorm (n = 40 , mean = 0 , sd = 1 )``` ```{r} set.seed (123 )<- replicate (n = 10000 , rnorm (40 ))hist (error_fixed, xlab = expression (hat (theta[k])~- ~ hat (theta)), prob = TRUE , breaks = 100 , ylim = c (0 , .45 ), xlim = c (- 4 ,4 ),main = "No Heterogeneity" )lines (seq (- 4 , 4 , 0.01 ), dnorm (seq (- 4 , 4 , 0.01 )), col = "blue" , lwd = 2 )``` ```{r} set.seed (123 )<- replicate (n = 10000 , rnorm (40 ) + rnorm (40 ))hist (error_random, xlab = expression (hat (theta[k])~- ~ hat (theta)), prob = TRUE , breaks = 100 ,ylim = c (0 , .45 ), xlim = c (- 4 ,4 ),main = "Heterogeneity" )lines (seq (- 4 , 4 , 0.01 ), dnorm (seq (- 4 , 4 , 0.01 )), col = "blue" , lwd = 2 )``` ```{r} set.seed (123 )<- replicate (10000 , sum (rnorm (40 )^ 2 ))<- replicate (10000 , sum ((rnorm (40 ) + rnorm (40 ))^ 2 ))``` ```{r} <- 40-1 hist (Q_fixed, xlab = expression (italic ("Q" )), prob = TRUE , breaks = 100 , ylim = c (0 , .06 ),xlim = c (0 ,160 ),main = "No Heterogeneity" )lines (seq (0 , 100 , 0.01 ), dchisq (seq (0 , 100 , 0.01 ), df = df), col = "blue" , lwd = 2 )hist (Q_random, xlab = expression (italic ("Q" )), prob = TRUE , breaks = 100 , ylim = c (0 , .06 ), xlim = c (0 ,160 ),main = "Heterogeneity" )lines (seq (0 , 100 , 0.01 ), dchisq (seq (0 , 100 , 0.01 ), df = df), col = "blue" , lwd = 2 )``` ## **Higgins & Thompson’s** $I^2$Statistic ```{r} # Display the value of the 10th simulation of Q 10 ]# Define k <- 40 # Calculate I^2 10 ] - (k-1 ))/ Q_fixed[10 ]``` ```{r} 10 ] - (k-1 ))/ Q_random[10 ]``` - I2 = 25%: low heterogeneity- I2 = 50%: moderate heterogeneity- I2 = 75%: substantial heterogeneity.## $H^2$ statistic ## **Heterogeneity Variance** $\tau^2$& Standard Deviation $\tau$ ```{r} library (meta)<- metagen (TE = TE,seTE = seTE,studlab = Author,data = dmetar:: ThirdWave,sm = "SMD" ,fixed = FALSE ,random = TRUE ,method.tau = "REML" ,method.random.ci = "HK" ,title = "Third Wave Psychotherapies" )``` ```{r} # Pooled effect $ TE.random# Estimate of tau $ tau``` ## Prediction intervals (PIs) 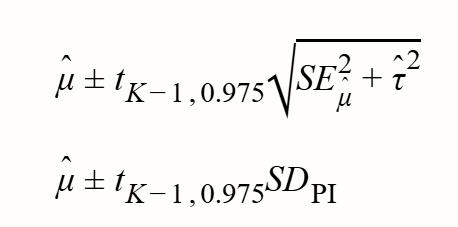 {fig-align="center"}```{r} <- update (m.gen, prediction = TRUE )summary (m.gen)``` ## **异常值** ### 删除 ```{r} :: find.outliers (m.gen)``` ### 影响分析 ```{r} <- dmetar:: InfluenceAnalysis (m.gen, random = TRUE )``` ```{r} plot (m.gen.inf, "baujat" )``` ```{r} plot (m.gen.inf, "influence" )``` ```{r} plot (m.gen.inf, "es" )plot (m.gen.inf, "i2" )``` ### **GOSH Plot Analysis** ```{r} library (metafor)<- rma (yi = m.gen$ TE,sei = m.gen$ seTE,method = m.gen$ method.tau,test = "knha" )``` ```{r eval=FALSE} res.gosh <- gosh(m.rma) save(res.gosh,file = "data/res.gosh.RData") ``` ```{r} load ("data/res.gosh.RData" )plot (res.gosh, alpha = 0.01 )``` ```{r} #' @usage gosh.diagnostics(data, km = TRUE, db = TRUE, gmm = TRUE, #' km.params = list(centers = 3, #' iter.max = 10, nstart = 1, #' algorithm = c("Hartigan-Wong", #' "Lloyd", "Forgy","MacQueen"), #' trace = FALSE), #' db.params = list(eps = 0.15, MinPts = 5, #' method = c("hybrid", "raw", "dist")), #' gmm.params = list(G = NULL, modelNames = NULL, #' prior = NULL, control = emControl(), #' initialization = list(hcPairs = NULL, #' subset = NULL, #' noise = NULL), #' Vinv = NULL, #' warn = mclust.options("warn"), #' x = NULL, verbose = FALSE), #' seed = 123, #' verbose = TRUE) ``` ```{r eval=FALSE} res.gosh.diag <- dmetar::gosh.diagnostics(res.gosh, km.params = list(centers = 2), db.params = list(eps = 0.08, MinPts = 50)) ``` ```{r} data ("m.gosh" ,package = "dmetar" )<- dmetar:: gosh.diagnostics (m.gosh)plot (res.diag)``` ```{r} update (m.gen, exclude = c (3 , 4 , 16 )) %>% summary ()```