7 方差分析
方差分析的假设
独立性
正态性
shapiro.test()方差齐性 正态
bartlett.test(),非正态Levene's test
7.1 单因素组间方差分析
7.1.1 计算公式
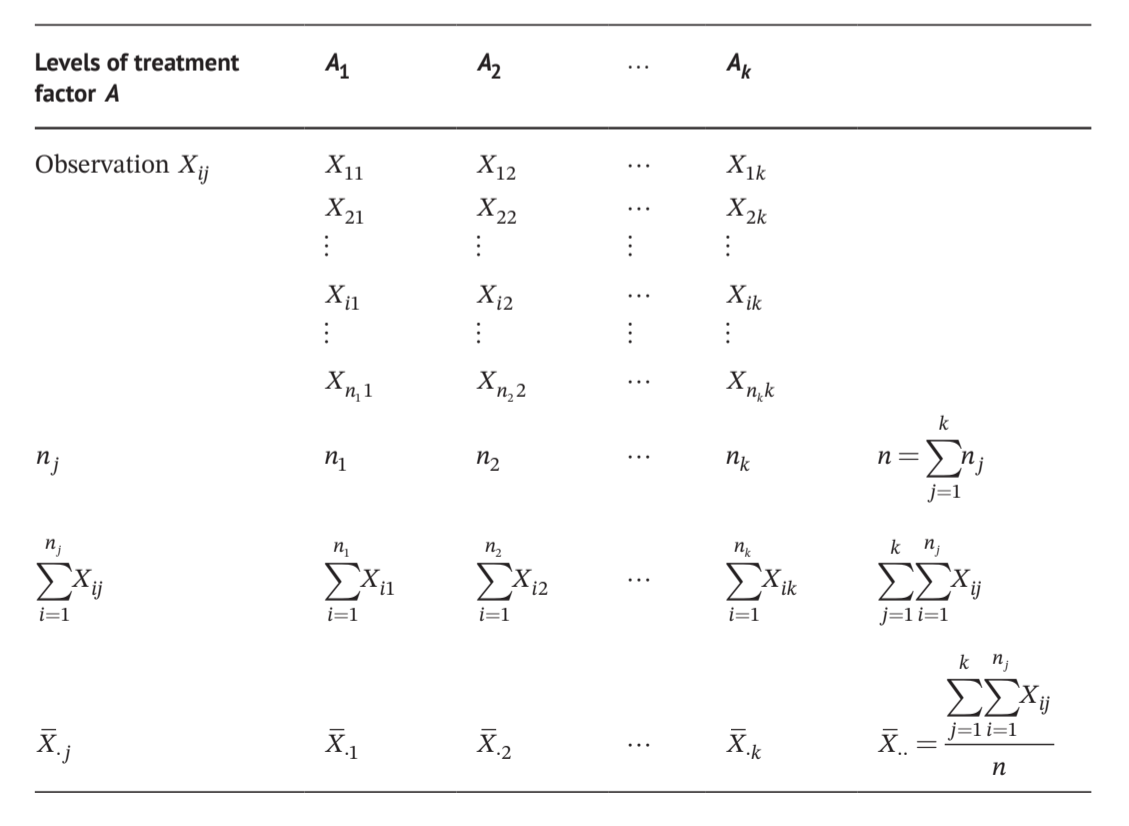
因子A有\(A_1,A_2,...,A_k\)共k个水平
total sum of squares
\[ SS_T=\sum_{j=1}^{k}\sum_{i=1}^{n_j}X_{ij}^2-\frac{(\sum_{j=1}^{k}\sum_{i=1}^{n_j}X_{ij})^2}{n}=SS_{组间}+SS_{组内} \]
between groups sum of squares \[ SS_{组间}=\sum_{j=1}^{k}\frac{(\sum_{i=1}^{n_j}X_{ij})^2}{n_j}-\frac{(\sum_{j=1}^{k}\sum_{i=1}^{n_j}X_{ij})^2}{n} \]
自由度\(\nu=n-1,\nu_{组间}=k-1,\nu_{组内}=\sum_{j=1}^{k}(n_j-1)=n-k\)
Between groups mean square \(MS_{组间}=\frac{SS_{组间}}{k-1}\)
Within groups mean square \(MS_{组内}=\frac{SS_{组内}}{n-k}\)
\[H_0:\mu_1=\mu_2=...=\mu_k\]
\[ \frac{SS_T}{\sigma^2}\sim \chi^2(\nu),\nu=n-1 \] \[ \frac{SS_{组内}}{\sigma^2}\sim \chi^2(\nu),\nu=n-k \] 因此,
\[ \frac{SS_{组间}}{\sigma^2}=\frac{SS_T}{\sigma^2}-\frac{SS_{组内}}{\sigma^2}\ \ \ \sim \chi^2(\nu),\nu=k-1 \]
检验统计量
\[ F=\frac{\frac{SS_{组间}}{(k-1)\sigma^2}}{\frac{SS_{组内}}{(n-k)\sigma^2}}=\frac{\frac{SS_{组间}}{k-1}}{\frac{SS_{组内}}{n-k}}=\frac{MS_{组间}}{MS_{组内}}\ \ \ \sim \chi^2(\nu),\nu=k-1 \]
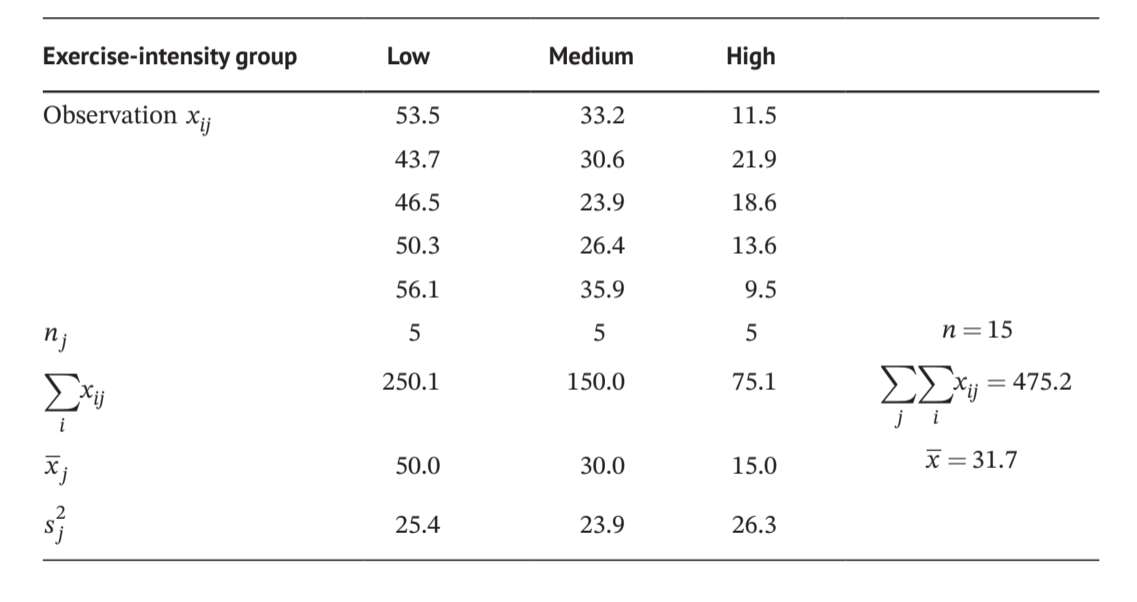
7.1.2 手算
Show the code
k <- 3
df_sum <- sum(df)
df_sum_square <- sum(df^2)
n <- 15
C <- df_sum^2/n
SS_T <- df_sum_square-C
SS_between <- apply(df, 2, function(x) sum(sum(x)^2/length(x))) |> sum()-C
SS_within <- SS_T-SS_between
MS_between <- SS_between/(k-1)
MS_within <- SS_within/(n-k)
F_stat <-MS_between/MS_within
p_value <- pf(F_stat,2,12,lower.tail = F)
7.1.3 stats::aov()
Show the code
df_long <- df |> pivot_longer(cols = everything(),
names_to = "level",
values_to = "value")
df_long$level <- factor(df_long$level)
df_aov <- aov(value~level,data = df_long)
anova(df_aov)
#> Analysis of Variance Table
#>
#> Response: value
#> Df Sum Sq Mean Sq F value Pr(>F)
#> level 2 3083.7 1541.83 61.245 5.046e-07 ***
#> Residuals 12 302.1 25.17
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
7.1.4 ez::ezANOVA()
Show the code
df_long <- df_long |> rowid_to_column(var = "id") |>
relocate(id,.before = 1) |> mutate(id=factor(id))
ez::ezANOVA(data = df_long,
dv = value,
wid = id,
between = level,
type = 3,
detailed = T)
#> $ANOVA
#> Effect DFn DFd SSn SSd F p p<.05 ges
#> 1 (Intercept) 1 12 15054.336 302.096 597.99545 1.318663e-11 * 0.9803277
#> 2 level 2 12 3083.668 302.096 61.24546 5.045797e-07 * 0.9107746
#>
#> $`Levene's Test for Homogeneity of Variance`
#> DFn DFd SSn SSd F p p<.05
#> 1 2 12 0.05733333 92.36 0.003724556 0.9962835
7.1.5 stats::lm()
Show the code
lm_aov <- lm(formula = value ~ level, data = df_long)
lm_aov
#>
#> Call:
#> lm(formula = value ~ level, data = df_long)
#>
#> Coefficients:
#> (Intercept) levellow levelmedium
#> 15.02 35.00 14.98
anova(lm_aov)
#> Analysis of Variance Table
#>
#> Response: value
#> Df Sum Sq Mean Sq F value Pr(>F)
#> level 2 3083.7 1541.83 61.245 5.046e-07 ***
#> Residuals 12 302.1 25.17
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
model.matrix(~ level, data = df_long)
#> (Intercept) levellow levelmedium
#> 1 1 1 0
#> 2 1 0 1
#> 3 1 0 0
#> 4 1 1 0
#> 5 1 0 1
#> 6 1 0 0
#> 7 1 1 0
#> 8 1 0 1
#> 9 1 0 0
#> 10 1 1 0
#> 11 1 0 1
#> 12 1 0 0
#> 13 1 1 0
#> 14 1 0 1
#> 15 1 0 0
#> attr(,"assign")
#> [1] 0 1 1
#> attr(,"contrasts")
#> attr(,"contrasts")$level
#> [1] "contr.treatment"
lm(formula = value ~ 0 + level, data = df_long)
#>
#> Call:
#> lm(formula = value ~ 0 + level, data = df_long)
#>
#> Coefficients:
#> levelhigh levellow levelmedium
#> 15.02 50.02 30.00
model.matrix(~ 0 + level, data = df_long)
#> levelhigh levellow levelmedium
#> 1 0 1 0
#> 2 0 0 1
#> 3 1 0 0
#> 4 0 1 0
#> 5 0 0 1
#> 6 1 0 0
#> 7 0 1 0
#> 8 0 0 1
#> 9 1 0 0
#> 10 0 1 0
#> 11 0 0 1
#> 12 1 0 0
#> 13 0 1 0
#> 14 0 0 1
#> 15 1 0 0
#> attr(,"assign")
#> [1] 1 1 1
#> attr(,"contrasts")
#> attr(,"contrasts")$level
#> [1] "contr.treatment"7.1.6 事后检验
成对比较的数量 \(N=\frac{k!}{2!(k-2)!},k≥3\),导致犯第Ⅰ类错误的概率迅速增加,\([1-(1-\alpha)^N]\)。
7.1.6.1 Tukey’s test
Tukey’s test 也被称为Tukey’s honestly significant difference (Tukey’s HSD) test。
- k个均值从大到小排列;
- 均值最大的组依次与均值最小,第二小,……,第二大比较;
- 均值第二大的组以同样的方式比较;
- 以此类推
- 在各组样本量相等的情况下,如果在两个均值之间未发现显著差异,则推断这两个均值所包含的任何均值之间不存在显著差异,并且不再检验所包含均值之间的差异。
studentized range statistic \(q=\frac{\bar X_{max}-\bar X_{min}}{S_{\bar X_{max}-\bar X_{min}}}\),其中\(S_{\bar X_{max}-\bar X_{min}}=\sqrt{\frac{MS_{组内}}{n}}\),\(n\)是每一个治疗组的样本量。
如果各组样本量不等,则\(S_{\bar X_{max}-\bar X_{min}}=\sqrt{\frac{MS_{组内}}{2}(\frac{1}{n_i}+\frac{1}{n_j})}\)
检验统计量
\[ HSD=q_{(k,\nu_{组内}),1-\alpha} \times S_{\bar X_{max}-\bar X_{min}},\nu_{组内}=k(n_j-1) \]
对于任意i,j且\(\bar X_i>\bar X_j\),如果\(\bar X_i-\bar X_j>HSD\),那么拒绝\(H_0\),说明这两组存在显著差异。
\[ H_0:\mu_i=\mu_j(i≠j) \]
Show the code
k_means <- apply(df,2,mean) |> sort(,decreasing = TRUE)
k_means
#> low medium high
#> 50.02 30.00 15.02
# 附录q q(3,12),1-0.05 3(5-1)=12
q_critical_value <- 3.77
nj <- 5
HSD <- q_critical_value*sqrt(MS_within/nj)
k_means
#> low medium high
#> 50.02 30.00 15.02
diff_means <- c()
names_diff_means <- c()
for(i in 1:(length(k_means)-1)){
for(j in length(k_means):(i+1)){
diff_value <- k_means[i] - k_means[j]
diff_means <- c(diff_means, diff_value)
names_diff_means <- c(names_diff_means, paste(names(k_means)[i], "vs.", names(k_means)[j],sep = "_"))
}
}
names(diff_means) <- names_diff_means
diff_means
#> low_vs._high low_vs._medium medium_vs._high
#> 35.00 20.02 14.98
#全为真,3组成对比较都存在显著差异
diff_means > HSD
#> low_vs._high low_vs._medium medium_vs._high
#> TRUE TRUE TRUEShow the code
pairwise <- TukeyHSD(df_aov)
pairwise
#> Tukey multiple comparisons of means
#> 95% family-wise confidence level
#>
#> Fit: aov(formula = value ~ level, data = df_long)
#>
#> $level
#> diff lwr upr p adj
#> low-high 35.00 26.534054 43.46595 0.0000003
#> medium-high 14.98 6.514054 23.44595 0.0013315
#> medium-low -20.02 -28.485946 -11.55405 0.00010697.1.6.2 Dunnett’s test
Dunnett’s test也称为q’-test,是两独立样本t-test的一种修正。Dunnett’s test 假设数据符合正态分布,并且各组的方差相等。 控制对照组(C)与其他每个实验组(T)比较。
\(H_0:\mu_C=\mu_T\)
\[ q'=\frac{\bar X_T-\bar X_C}{\sqrt{MS_{组内}(\frac{1}{n_T}+\frac{1}{n_C})}} \sim q'(\nu,a) \ \ \nu=\nu_{组内},a=k \]
临界值 \(q'_{(a,\nu_E),1-\alpha/2}\)
Show the code
nT <- nC <- 5
SE_mean_diff <- sqrt(MS_within*(1/nT+1/nC))
k_means
#> low medium high
#> 50.02 30.00 15.02
diff_means <- c()
names_diff_means <- c()
# 以 low 作控制组
for(i in 2:length(k_means)){
diff_value <- k_means[i] - k_means[1]
diff_means <- c(diff_means, diff_value)
names_diff_means <- c(names_diff_means, paste(names(k_means)[1], "vs.", names(k_means)[i],sep = "_"))
}
names(diff_means) <- names_diff_means
diff_means
#> low_vs._medium low_vs._high
#> -20.02 -35.00
q撇_stat <- diff_means/SE_mean_diff
# 附录q' q'(3,12),1-0.05/2
abs(q撇_stat)>2.50
#> low_vs._medium low_vs._high
#> TRUE TRUE
#全为真,各实验组与控制组均存在显著差异Show the code
library(multcomp)
dunnett_result <- glht(df_aov, linfct =mcp(level =c("medium - low = 0", "high - low = 0")))
# 查看 Dunnett's test 结果
summary(dunnett_result)
#>
#> Simultaneous Tests for General Linear Hypotheses
#>
#> Multiple Comparisons of Means: User-defined Contrasts
#>
#>
#> Fit: aov(formula = value ~ level, data = df_long)
#>
#> Linear Hypotheses:
#> Estimate Std. Error t value Pr(>|t|)
#> medium - low == 0 -20.020 3.173 -6.309 7.46e-05 ***
#> high - low == 0 -35.000 3.173 -11.030 2.38e-07 ***
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#> (Adjusted p values reported -- single-step method)7.1.6.3 LSD-t test
least significant difference t-test
挑选任意感兴趣的两组进行比较
\[ H_0:\mu_i=\mu_j(i≠j) \]
\[ LSD-t=\frac{\bar X_i-\bar X_j}{\sqrt{MS_{组内}(\frac{1}{n_i}+\frac{1}{n_j})}} \sim t(\nu) \ ,\ \nu=\nu_{组内}=n-k,a=k \]
Show the code
# medium high
LSD <- (k_means[2]-k_means[3])/sqrt(MS_within*(1/5+1/5))
# 附录 t(12,1-0.05/2)=2.7197.2 方差分析假设
https://www.statmethods.net/stats/rdiagnostics.html
7.2.1 独立性
7.2.2 正态性
Show the code
# 异常观测点
car::outlierTest(df_aov)
#> No Studentized residuals with Bonferroni p < 0.05
#> Largest |rstudent|:
#> rstudent unadjusted p-value Bonferroni p
#> 6 1.63682 0.12993 NA7.2.2.1 直方图/茎叶图
7.2.2.2 P-P图/Q-Q图
7.2.2.3 偏度和峰度
\[ H_0:总体偏度系数\gamma_1=0 或者总体峰度系数\gamma_2=0 \]
\[ z_i=\frac{g_i-0}{\sigma_{g_i}} \ \ \ \ 临界值z_{1-\alpha/2} \]
7.2.2.4 Shapiro-Wilk检验(小样本)
Show the code
shapiro.test(df$low)
#>
#> Shapiro-Wilk normality test
#>
#> data: df$low
#> W = 0.9718, p-value = 0.8867
shapiro.test(df$medium)
#>
#> Shapiro-Wilk normality test
#>
#> data: df$medium
#> W = 0.96762, p-value = 0.8598
shapiro.test(df$high)
#>
#> Shapiro-Wilk normality test
#>
#> data: df$high
#> W = 0.94361, p-value = 0.6916
# 因为观测太少,也可以同时检验
shapiro.test(c(df$low,df$medium,df$high))
#>
#> Shapiro-Wilk normality test
#>
#> data: c(df$low, df$medium, df$high)
#> W = 0.94559, p-value = 0.45787.2.2.5 Kolmogorov-Smirnov检验(Lilliefors correction 大样本)
Show the code
ks.test(c(df$low,df$medium,df$high),"pnorm")
#>
#> Exact one-sample Kolmogorov-Smirnov test
#>
#> data: c(df$low, df$medium, df$high)
#> D = 1, p-value < 2.2e-16
#> alternative hypothesis: two-sided
ks.test(rnorm(1000),"pnorm")
#>
#> Asymptotic one-sample Kolmogorov-Smirnov test
#>
#> data: rnorm(1000)
#> D = 0.036487, p-value = 0.1395
#> alternative hypothesis: two-sidedShow the code
x <- rnorm(50)
y <- runif(50)
ks.test(x, y) # perform ks test
#>
#> Exact two-sample Kolmogorov-Smirnov test
#>
#> data: x and y
#> D = 0.46, p-value = 3.801e-05
#> alternative hypothesis: two-sided
x <- rnorm(50)
y <- rnorm(50)
ks.test(x, y)
#>
#> Exact two-sample Kolmogorov-Smirnov test
#>
#> data: x and y
#> D = 0.1, p-value = 0.9667
#> alternative hypothesis: two-sided7.2.3 方差齐性
http://www.cookbook-r.com/Statistical_analysis/Homogeneity_of_variance/
7.2.3.1 Bartlett’s test
数据满足正态性
比较每一组方差的加权算术均值和几何均值。
\[ H_0:\sigma_1^2=\sigma_2^2=...=\sigma_k^2 \] 当样本量\(n_j\)≥5时,检验统计量(各组样本量相等)
\[ B=\frac{(n-1)[kln\bar S^2-\sum_{j=1}^{k}lnS_j^2]}{1+\frac{k+1}{3k(n-1)}} \sim\ \chi^2(\nu)\ ,\nu=k-1 \] 其中n是每一组的样本量,\(S_j^2\)是某一组的样本方差,\(\bar S^2\)是所有k个组样本方差的平均值。
当各组样本量不等时,
\[ B=\frac{\sum_{j=1}^{k}(n_j-1)ln\frac{\bar S^2}{S_j^2}}{1+\frac{1}{3(k-1)}(\sum_{j=1}^{k}\frac {1}{n_j-1}-\frac{1}{\sum_{j=1}^{k}(n_j-1)})} \sim\ \chi^2(\nu)\ ,\nu=k-1 \] 其中\(h_j\)是某一组的样本量,\(\bar S^2=(\sum_{j=1}^{k}(n_j-1)S_j^2)/(\sum_{j=1}^{k}(n_j-1))\)是所有k个组样本方差的加权平均值。
Show the code
Show the code
bartlett.test(df)
#>
#> Bartlett test of homogeneity of variances
#>
#> data: df
#> Bartlett's K-squared = 0.0081595, df = 2, p-value = 0.99597.2.3.2 Levene’s test
数据不满足正态性
- k个随机样本是独立的
- 随机变量X是连续的
Levene 变换:
\[ Z_{ij}=|X_{ij}-\bar X_{.\ j}|(i=1,2...,n_j;\ j=1,2,...,k) \]
检验统计量(基于变换后的F检验)
\[ W=\frac{MS_{组间}}{MS_{组内}}=\frac{\sum_jn_j(\bar Z_{.j}-\bar Z_{..})^2/(k-1)}{\sum_j\sum_i(Z_{ij}-\bar Z_{.j})^2/(n-k)} \sim F(\nu_1,\nu_2) \ \ \ \ \nu_1=k-1,\nu_2=n-k \]
Show the code
v1=3-1
v2=15-3
z.j_mean <- apply(z_df, 2, mean)
z.j_mean
#> low medium high
#> 3.9 3.9 4.2
z_mean <- mean(z.j_mean)
z_mean
#> [1] 4
options(digits = 4)
W <- 12*sum(5*(z.j_mean-z_mean)^2)/(2*(sum((z_df$low-z.j_mean[1])^2)+sum((z_df$medium-z.j_mean[2])^2)+sum((z_df$high-z.j_mean[3])^2)))
W
#> [1] 0.02547.3 Welch’s ANOVA Test
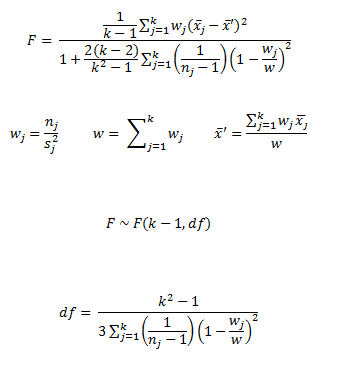
7.4 数据变换
当方差分析的正态性假设或方差齐性假设不为真时,通常使用(1)数据变换方法;(2)非参数检验方法 比较均值差异
7.4.1 平方根变换
当每个水平组的方差与均值成比例,尤其是样本来自泊松分布
\[ Y=\sqrt{X} \]
当数据中有零或非常小的值时,
\[ Y=\sqrt{X+a} \ \ \ \ a=0.5或0.1 \]
7.4.2 对数变换
当数据方差不齐且每个水平组标准差与均值成比例时
\[ Y=\log{X} \ \ \ \ base=e或10 \]
当数据中有零或负值时,
\[ Y=\log{(X+a)} \ \ \ \ a为实数,使得X+a>0 \]
7.4.3 反正弦平方根变换
率,服从二项分布\(B(n,\pi)\)
\[ Y=\arcsin {\sqrt{\pi}} \ \ \ \ \]