线性混合效应模型 (Linear Mixed-effect Model, LMM)用于分析具有固定效应和随机效应的数据结构。它能够处理数据中的组内相关性和个体差异,广泛应用于生态学、心理学等领域的统计分析。如果数据满足正态分布但有组内相关性,使用线性混合模型(LMM)。
Mixed Effects Models and Extensions in Ecology with R 第5章
线性混合模型的一般表达形式:
\[
Y = \mathbf{X} \beta + \mathbf{Z} \gamma + \epsilon
\]
其中:
lme4::lmer()
lmer()
\[
lmer (data,formual= DV \sim Fixed\_Factor + (Random\_intercept + Random\_slope | Random\_Factor))
\]
截距中,1表示随机截距,0表示固定截距,默认截距为1。
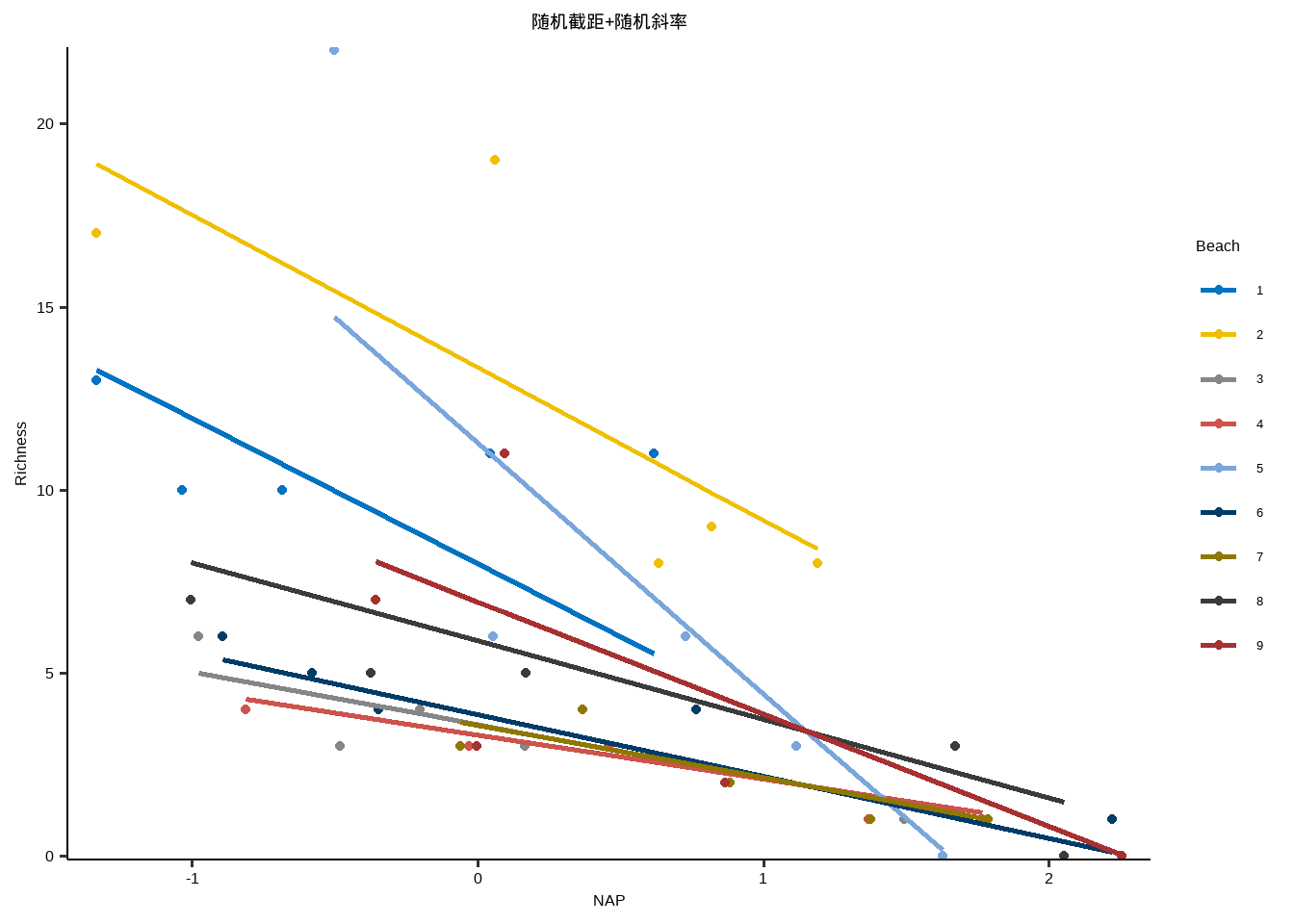
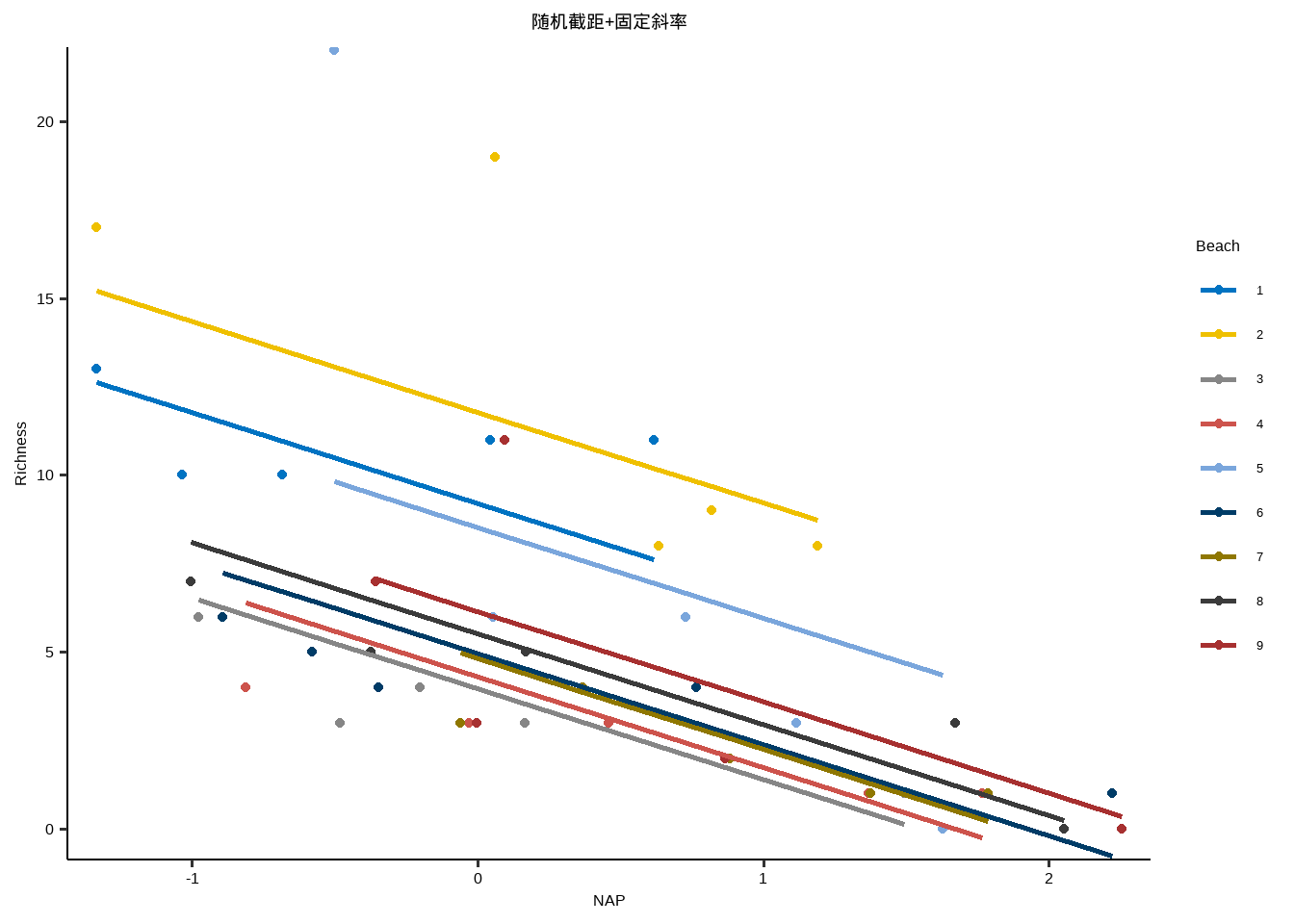
随机截距+随机斜率
y~x+( 1+x | id )
y~x+( x | id )
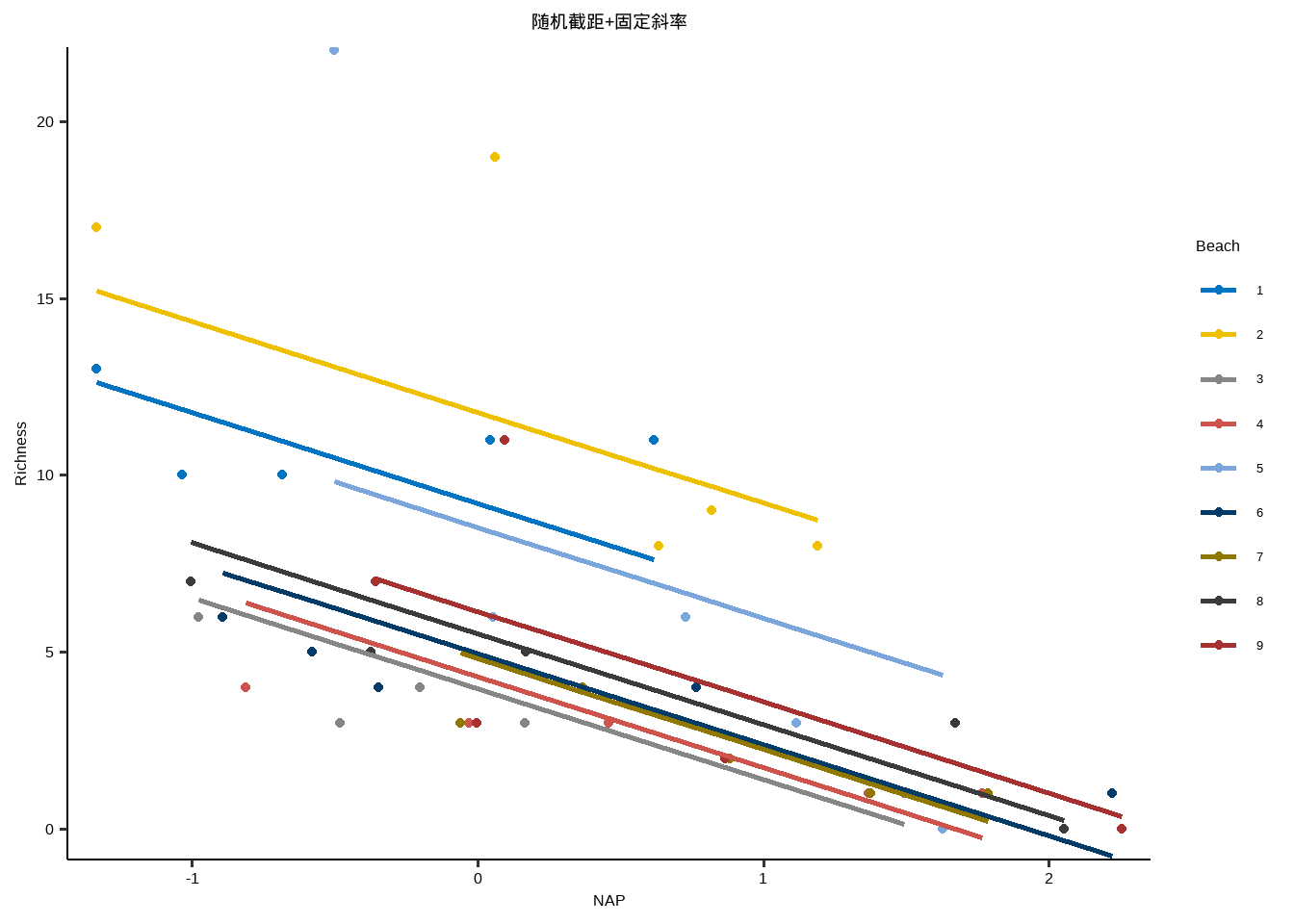
随机截距+固定斜率
y~x+( 1+1 | id )
y~x+( 1 | id )
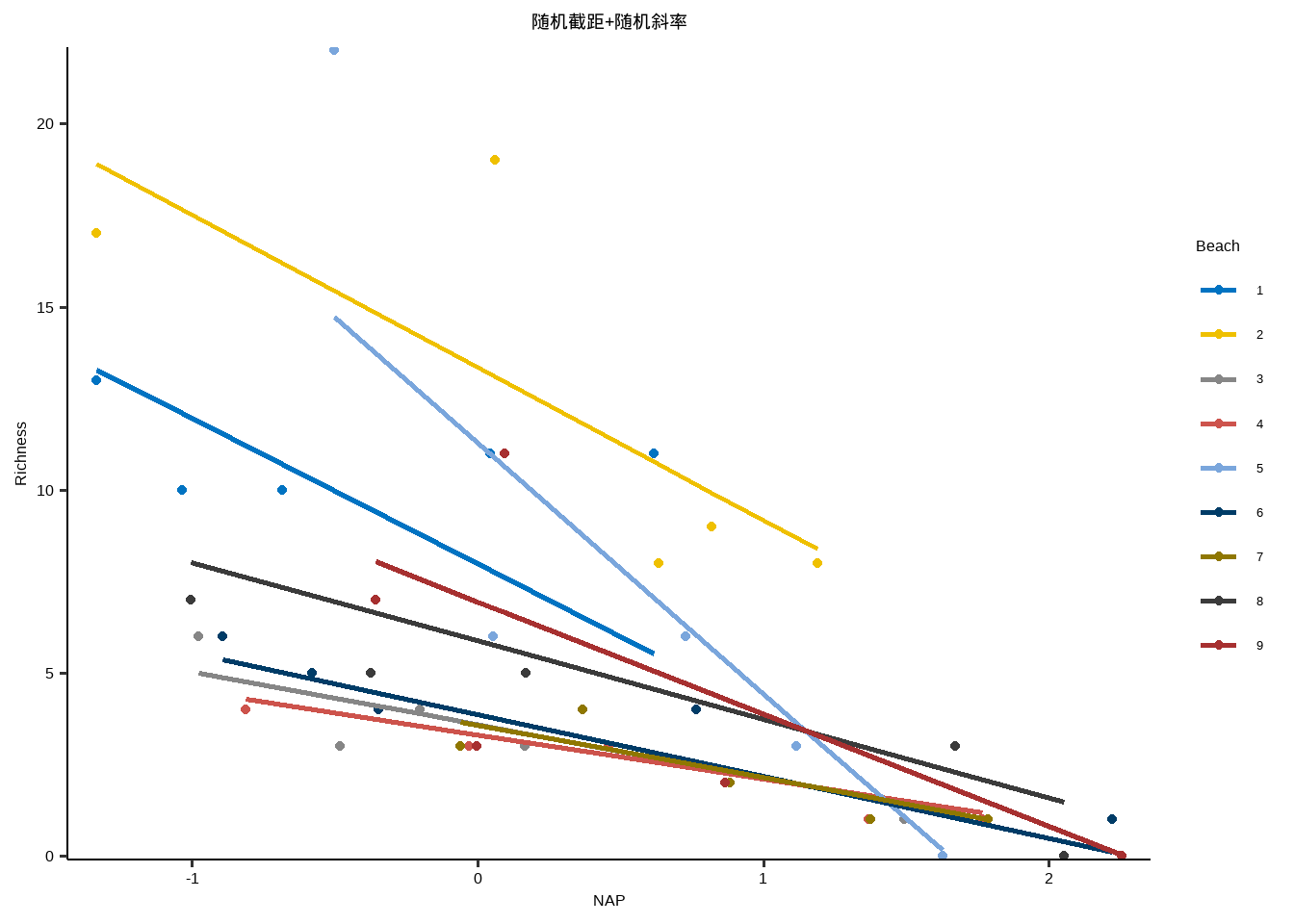
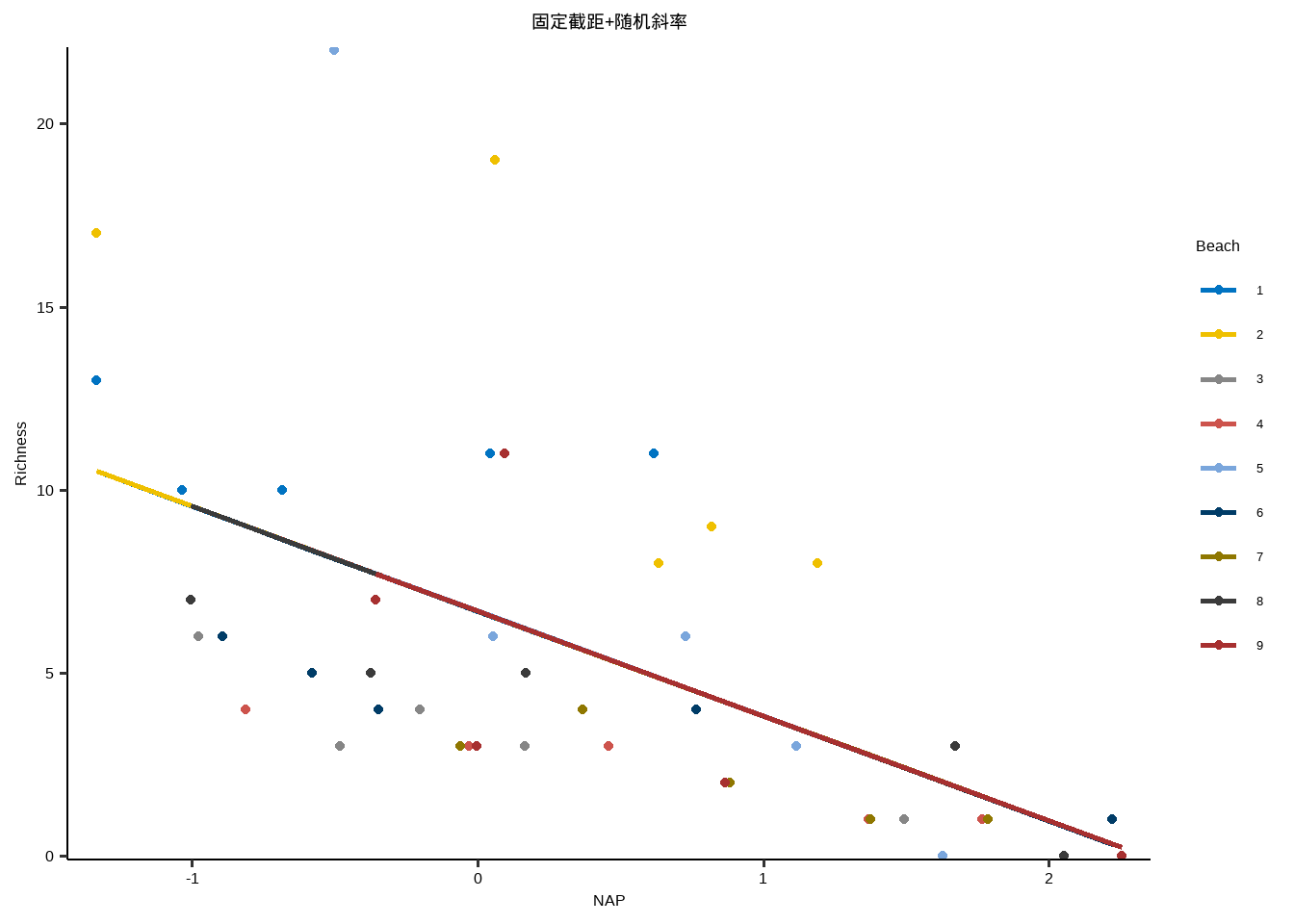
固定截距+随机斜率
y~x+( 0+x | id )
NA
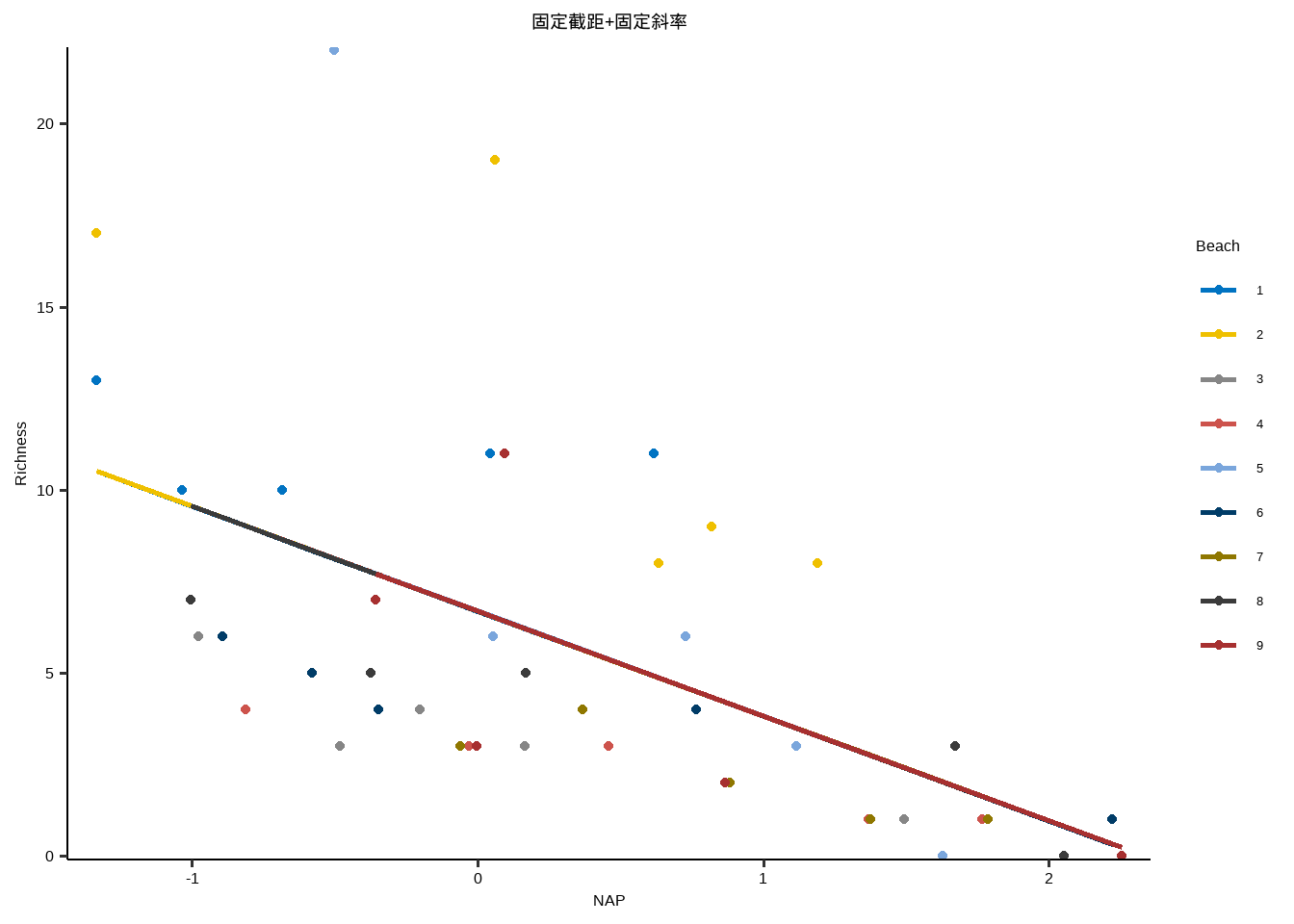
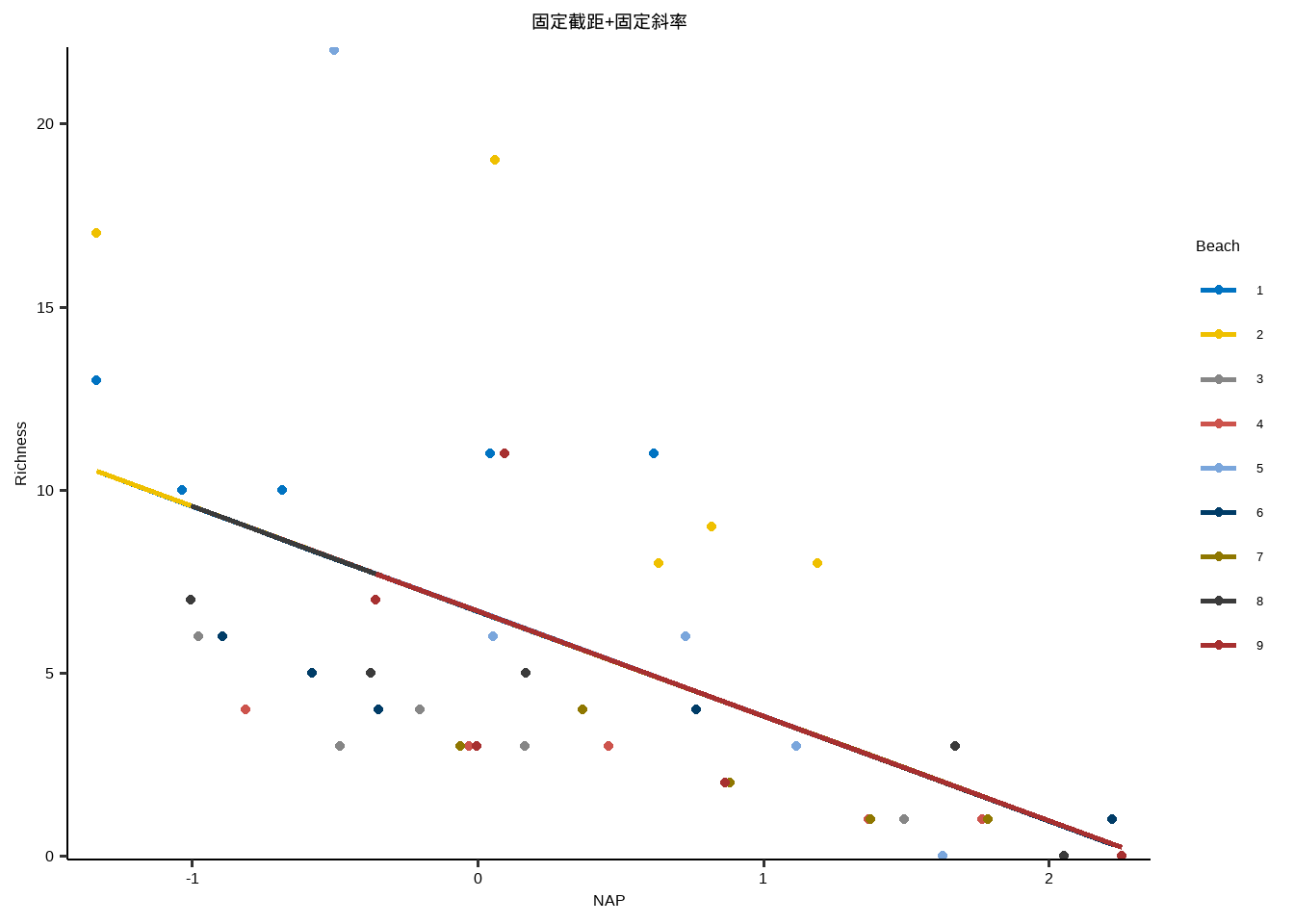
线性模型:固定截距+固定斜率
y~x
NA
随机截距+随机斜率
Show the code df_long <- read_delim ( "data/AED/RIKZ.txt" ) df_long $ Beach <- factor ( df_long $ Beach ) df_long $ Exposure <- factor ( df_long $ Exposure ) head ( df_long ) #> # A tibble: 6 × 5 #> Sample Richness Exposure NAP Beach #> <dbl> <dbl> <fct> <dbl> <fct> #> 1 1 11 10 0.045 1 #> 2 2 10 10 -1.04 1 #> 3 3 13 10 -1.34 1 #> 4 4 11 10 0.616 1 #> 5 5 10 10 -0.684 1 #> 6 6 8 8 1.19 2
\[
\eta_{(nrow \times 1)} = \mathbf{X}_{nrow \times 1} \beta_{1 \times 1} + \mathbf{Z}_{nrow \times 2n_{subjects}} \mathbf{\gamma}_{2n_{subjects} \times 1} + \epsilon_i
\]
Z有两倍于受试者数量的列,每个受试者的随机截距和随机斜率
Show the code library ( lme4 ) lme1 <- lmer ( Richness ~ 1 + NAP * Exposure + ( 1 + NAP | Beach ) ,data = df_long ) summary ( lme1 ) #> Linear mixed model fit by REML ['lmerMod'] #> Formula: Richness ~ 1 + NAP * Exposure + (1 + NAP | Beach) #> Data: df_long #> #> REML criterion at convergence: 207.2 #> #> Scaled residuals: #> Min 1Q Median 3Q Max #> -1.92384 -0.36066 -0.13343 0.09819 2.84228 #> #> Random effects: #> Groups Name Variance Std.Dev. Corr #> Beach (Intercept) 3.758 1.938 #> NAP 2.837 1.684 -1.00 #> Residual 6.535 2.556 #> Number of obs: 45, groups: Beach, 9 #> #> Fixed effects: #> Estimate Std. Error t value #> (Intercept) 13.3457 2.2784 5.858 #> NAP -4.1753 2.1243 -1.965 #> Exposure10 -5.3273 2.5556 -2.085 #> Exposure11 -9.7660 2.5653 -3.807 #> NAP:Exposure10 0.1646 2.3621 0.070 #> NAP:Exposure11 2.7273 2.3715 1.150 #> #> Correlation of Fixed Effects: #> (Intr) NAP Exps10 Exps11 NAP:E10 #> NAP -0.770 #> Exposure10 -0.892 0.686 #> Exposure11 -0.888 0.683 0.792 #> NAP:Expsr10 0.692 -0.899 -0.775 -0.615 #> NAP:Expsr11 0.689 -0.896 -0.615 -0.779 0.806 #> optimizer (nloptwrap) convergence code: 0 (OK) #> boundary (singular) fit: see help('isSingular') AIC ( lme1 ) #> [1] 227.1558 BIC ( lme1 ) #> [1] 245.2224 logLik ( lme1 ) #> 'log Lik.' -103.5779 (df=10) 2 * ( 1 - pt ( - 3.914605 ,35 ,lower.tail = F ) ) #> [1] 0.0003993968 library ( broom.mixed ) tidy ( lme1 ) #> # A tibble: 10 × 6 #> effect group term estimate std.error statistic #> <chr> <chr> <chr> <dbl> <dbl> <dbl> #> 1 fixed <NA> (Intercept) 13.3 2.28 5.86 #> 2 fixed <NA> NAP -4.18 2.12 -1.97 #> 3 fixed <NA> Exposure10 -5.33 2.56 -2.08 #> 4 fixed <NA> Exposure11 -9.77 2.57 -3.81 #> 5 fixed <NA> NAP:Exposure10 0.165 2.36 0.0697 #> 6 fixed <NA> NAP:Exposure11 2.73 2.37 1.15 #> 7 ran_pars Beach sd__(Intercept) 1.94 NA NA #> 8 ran_pars Beach cor__(Intercept).NAP -1 NA NA #> 9 ran_pars Beach sd__NAP 1.68 NA NA #> 10 ran_pars Residual sd__Observation 2.56 NA NA library ( nlme ) nlme1 <- lme ( Richness ~ 1 + NAP * Exposure , random = ~ 1 + NAP | Beach ,
data = df_long ,
control = lmeControl ( opt = "optim" , msMaxIter = 100 , msMaxEval = 5000 ) )
summary ( nlme1 ) #> Linear mixed-effects model fit by REML #> Data: df_long #> AIC BIC logLik #> 227.2046 243.8402 -103.6023 #> #> Random effects: #> Formula: ~1 + NAP | Beach #> Structure: General positive-definite, Log-Cholesky parametrization #> StdDev Corr #> (Intercept) 1.939709 (Intr) #> NAP 1.689580 -0.996 #> Residual 2.554676 #> #> Fixed effects: Richness ~ 1 + NAP * Exposure #> Value Std.Error DF t-value p-value #> (Intercept) 13.345694 2.278989 33 5.855970 0.0000 #> NAP -4.175271 2.128084 33 -1.961986 0.0582 #> Exposure10 -5.323695 2.556406 6 -2.082492 0.0825 #> Exposure11 -9.765613 2.566035 6 -3.805720 0.0089 #> NAP:Exposure10 0.167146 2.366467 33 0.070631 0.9441 #> NAP:Exposure11 2.726865 2.375786 33 1.147774 0.2593 #> Correlation: #> (Intr) NAP Exps10 Exps11 NAP:E10 #> NAP -0.767 #> Exposure10 -0.891 0.684 #> Exposure11 -0.888 0.682 0.792 #> NAP:Exposure10 0.690 -0.899 -0.772 -0.613 #> NAP:Exposure11 0.687 -0.896 -0.613 -0.777 0.806 #> #> Standardized Within-Group Residuals: #> Min Q1 Med Q3 Max #> -1.9251786 -0.3608959 -0.1327685 0.0992999 2.8420571 #> #> Number of Observations: 45 #> Number of Groups: 9 tidy ( nlme1 ) #> # A tibble: 10 × 8 #> effect group term estimate std.error df statistic p.value #> <chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> #> 1 fixed <NA> (Intercept) 13.3 2.28 33 5.86 1.47e-6 #> 2 fixed <NA> NAP -4.18 2.13 33 -1.96 5.82e-2 #> 3 fixed <NA> Exposure10 -5.32 2.56 6 -2.08 8.25e-2 #> 4 fixed <NA> Exposure11 -9.77 2.57 6 -3.81 8.91e-3 #> 5 fixed <NA> NAP:Exposure10 0.167 2.37 33 0.0706 9.44e-1 #> 6 fixed <NA> NAP:Exposure11 2.73 2.38 33 1.15 2.59e-1 #> 7 ran_pars Beach sd_(Intercept) 1.94 NA NA NA NA #> 8 ran_pars Beach cor_NAP.(Inter… -0.996 NA NA NA NA #> 9 ran_pars Beach sd_NAP 1.69 NA NA NA NA #> 10 ran_pars Residual sd_Observation 2.55 NA NA NA NA
这个模型的公式可以分解为:
Show the code # 标准化模型残差分布 quantile ( residuals ( lme1 ,type= "pearson" ,scaled= T ) ) #> 0% 25% 50% 75% 100% #> -1.9238372 -0.3606612 -0.1334336 0.0981939 2.8422803 # 随机因子随机效应和显著性检验 ranef ( lme1 ) #> $Beach #> (Intercept) NAP #> 1 -4.274356e-02 3.713708e-02 #> 2 -2.709479e-14 2.354089e-14 #> 3 3.819348e-03 -3.318381e-03 #> 4 -2.741786e-01 2.382158e-01 #> 5 3.269522e+00 -2.840673e+00 #> 6 2.736093e-01 -2.377212e-01 #> 7 -3.250033e-03 2.823741e-03 #> 8 -2.148947e+00 1.867079e+00 #> 9 -1.077831e+00 9.364564e-01 #> #> with conditional variances for "Beach" lmerTest :: ranova ( lme1 ) #> ANOVA-like table for random-effects: Single term deletions #> #> Model: #> Richness ~ NAP + Exposure + (1 + NAP | Beach) + NAP:Exposure #> npar logLik AIC LRT Df Pr(>Chisq) #> <none> 10 -103.58 227.16 #> NAP in (1 + NAP | Beach) 8 -105.67 227.35 4.1935 2 0.1229 # 查看固定效应和显著性检验 coef ( lme1 ) #> $Beach #> (Intercept) NAP Exposure10 Exposure11 NAP:Exposure10 NAP:Exposure11 #> 1 13.30295 -4.138134 -5.327265 -9.766048 0.1646123 2.727307 #> 2 13.34569 -4.175271 -5.327265 -9.766048 0.1646123 2.727307 #> 3 13.34951 -4.178590 -5.327265 -9.766048 0.1646123 2.727307 #> 4 13.07152 -3.937055 -5.327265 -9.766048 0.1646123 2.727307 #> 5 16.61522 -7.015944 -5.327265 -9.766048 0.1646123 2.727307 #> 6 13.61930 -4.412992 -5.327265 -9.766048 0.1646123 2.727307 #> 7 13.34244 -4.172447 -5.327265 -9.766048 0.1646123 2.727307 #> 8 11.19675 -2.308192 -5.327265 -9.766048 0.1646123 2.727307 #> 9 12.26786 -3.238815 -5.327265 -9.766048 0.1646123 2.727307 #> #> attr(,"class") #> [1] "coef.mer" anova ( lme1 ) #> Analysis of Variance Table #> npar Sum Sq Mean Sq F value #> NAP 1 124.279 124.279 19.0178 #> Exposure 2 142.983 71.492 10.9400 #> NAP:Exposure 2 22.308 11.154 1.7068 # 查看 类和方法 class ( lme1 ) #> [1] "lmerMod" #> attr(,"package") #> [1] "lme4" # methods(class = "lmerMod") # confint(lme1,level = 0.95)
随机截距+固定斜率
\[
\eta_{(nrow \ \times 1)}=\mathbf{X_{nrow×1}}\beta_{1\times1} +Z_{nrow\times n_{subjects}} \mathbf{\gamma}_{n_{subjects}\times 1}+\epsilon_i
\]
Z有一倍于受试者数量的列,每个受试者的随机截距。
Show the code lme2 <- lmer ( Richness ~ 1 + NAP + ( 1 | Beach ) ,data = df_long ) summary ( lme2 ) #> Linear mixed model fit by REML ['lmerMod'] #> Formula: Richness ~ 1 + NAP + (1 | Beach) #> Data: df_long #> #> REML criterion at convergence: 239.5 #> #> Scaled residuals: #> Min 1Q Median 3Q Max #> -1.4227 -0.4848 -0.1576 0.2519 3.9794 #> #> Random effects: #> Groups Name Variance Std.Dev. #> Beach (Intercept) 8.668 2.944 #> Residual 9.362 3.060 #> Number of obs: 45, groups: Beach, 9 #> #> Fixed effects: #> Estimate Std. Error t value #> (Intercept) 6.5819 1.0958 6.007 #> NAP -2.5684 0.4947 -5.192 #> #> Correlation of Fixed Effects: #> (Intr) #> NAP -0.157 AIC ( lme2 ) #> [1] 247.4802 BIC ( lme2 ) #> [1] 254.7069 logLik ( lme2 ) #> 'log Lik.' -119.7401 (df=4) 2 * ( 1 - pt ( 6.007 ,35 ,lower.tail = T ) ) #> [1] 7.558855e-07 nlme2 <- lme ( Richness ~ 1 + NAP * Exposure ,random= ~ 1 | Beach ,data = df_long ) summary ( nlme2 ) #> Linear mixed-effects model fit by REML #> Data: df_long #> AIC BIC logLik #> 227.3493 240.6578 -105.6747 #> #> Random effects: #> Formula: ~1 | Beach #> (Intercept) Residual #> StdDev: 0.5138683 2.971793 #> #> Fixed effects: Richness ~ 1 + NAP * Exposure #> Value Std.Error DF t-value p-value #> (Intercept) 13.345694 1.483557 33 8.995739 0.0000 #> NAP -4.175271 1.505110 33 -2.774063 0.0090 #> Exposure10 -5.544983 1.657659 6 -3.345069 0.0155 #> Exposure11 -9.730595 1.670518 6 -5.824898 0.0011 #> NAP:Exposure10 0.671731 1.643864 33 0.408629 0.6855 #> NAP:Exposure11 2.688806 1.656743 33 1.622947 0.1141 #> Correlation: #> (Intr) NAP Exps10 Exps11 NAP:E10 #> NAP -0.278 #> Exposure10 -0.895 0.249 #> Exposure11 -0.888 0.247 0.795 #> NAP:Exposure10 0.255 -0.916 -0.276 -0.226 #> NAP:Exposure11 0.253 -0.908 -0.226 -0.296 0.832 #> #> Standardized Within-Group Residuals: #> Min Q1 Med Q3 Max #> -1.5652904 -0.4386841 -0.1164805 0.1783113 4.1098230 #> #> Number of Observations: 45 #> Number of Groups: 9
固定截距+随机斜率
\[
\eta_{(nrow \times 1)} = \mathbf{X}_{nrow \times 1} \beta_{1 \times 1} + \mathbf{Z}_{nrow \times n_{subjects}} \mathbf{\gamma}_{n_{subjects} \times 1} + \epsilon_i
\]
Z 有一倍于受试者数量的列,每个受试者的随机斜率。
Show the code lme3 <- lmer ( Richness ~ 1 + NAP + ( 0 + NAP | Beach ) ,data = df_long ) summary ( lme3 ) #> Linear mixed model fit by REML ['lmerMod'] #> Formula: Richness ~ 1 + NAP + (0 + NAP | Beach) #> Data: df_long #> #> REML criterion at convergence: 252.2 #> #> Scaled residuals: #> Min 1Q Median 3Q Max #> -1.2182 -0.6636 -0.1930 0.3253 3.3347 #> #> Random effects: #> Groups Name Variance Std.Dev. #> Beach NAP 0.00 0.00 #> Residual 17.31 4.16 #> Number of obs: 45, groups: Beach, 9 #> #> Fixed effects: #> Estimate Std. Error t value #> (Intercept) 6.6857 0.6578 10.164 #> NAP -2.8669 0.6307 -4.545 #> #> Correlation of Fixed Effects: #> (Intr) #> NAP -0.333 #> optimizer (nloptwrap) convergence code: 0 (OK) #> boundary (singular) fit: see help('isSingular') nlme3 <- lme ( Richness ~ 1 + NAP ,random= ~ 0 + NAP | Beach ,data = df_long ) summary ( nlme3 ) #> Linear mixed-effects model fit by REML #> Data: df_long #> AIC BIC logLik #> 260.201 267.2458 -126.1005 #> #> Random effects: #> Formula: ~0 + NAP | Beach #> NAP Residual #> StdDev: 0.0001127408 4.159929 #> #> Fixed effects: Richness ~ 1 + NAP #> Value Std.Error DF t-value p-value #> (Intercept) 6.685662 0.6577579 35 10.164320 0e+00 #> NAP -2.866853 0.6307186 35 -4.545376 1e-04 #> Correlation: #> (Intr) #> NAP -0.333 #> #> Standardized Within-Group Residuals: #> Min Q1 Med Q3 Max #> -1.2181663 -0.6636488 -0.1930031 0.3253447 3.3347473 #> #> Number of Observations: 45 #> Number of Groups: 9
随机效应模型
Show the code lme4 <- lmer ( Richness ~ 1 + ( 1 | Beach ) ,data = df_long ) summary ( lme4 ) #> Linear mixed model fit by REML ['lmerMod'] #> Formula: Richness ~ 1 + (1 | Beach) #> Data: df_long #> #> REML criterion at convergence: 261.1 #> #> Scaled residuals: #> Min 1Q Median 3Q Max #> -1.7797 -0.5070 -0.0980 0.2547 3.8063 #> #> Random effects: #> Groups Name Variance Std.Dev. #> Beach (Intercept) 10.48 3.237 #> Residual 15.51 3.938 #> Number of obs: 45, groups: Beach, 9 #> #> Fixed effects: #> Estimate Std. Error t value #> (Intercept) 5.689 1.228 4.631 nlme4 <- lme ( Richness ~ 1 ,random= ~ 1 | Beach ,data = df_long ) summary ( nlme4 ) #> Linear mixed-effects model fit by REML #> Data: df_long #> AIC BIC logLik #> 267.1142 272.4668 -130.5571 #> #> Random effects: #> Formula: ~1 | Beach #> (Intercept) Residual #> StdDev: 3.237112 3.938415 #> #> Fixed effects: Richness ~ 1 #> Value Std.Error DF t-value p-value #> (Intercept) 5.688889 1.228419 36 4.631066 0 #> #> Standardized Within-Group Residuals: #> Min Q1 Med Q3 Max #> -1.77968689 -0.50704111 -0.09795286 0.25468670 3.80631705 #> #> Number of Observations: 45 #> Number of Groups: 9
线性模型:固定截距+ 固定斜率
Show the code lm <- lm ( Richness ~ 1 + NAP ,data = df_long ) lm #> #> Call: #> lm(formula = Richness ~ 1 + NAP, data = df_long) #> #> Coefficients: #> (Intercept) NAP #> 6.686 -2.867
模型选择
限制最大似然法REML
赤池信息准则(AIC)
\[ kIC=−2log(\mathcal{L})+2k\]
其中:
\(\mathcal{L}\) 是似然函数。
\(k\) 是模型参数的数量。
贝叶斯信息准则(BIC)
\[ BIC=−2log(\mathcal{L})+klog(n) \]
其中:
Show the code plot_lme <- function ( model , title ) { ggplot ( df_long , aes ( NAP , Richness , group = Beach , color = Beach ) ) +
geom_point ( ) +
geom_line (
data = bind_cols ( df_long , .pred = predict ( model , df_long ) ) ,
mapping = aes ( y = .pred ) ,
linewidth = 1
) +
labs ( title = title ) +
scale_x_continuous ( expand = ( mult = c ( 0 ,.1 ) ) ) +
scale_y_continuous ( expand = ( mult = c ( 0 ,.1 ) ) ) +
ggsci :: scale_color_jco ( ) +
ggpubr :: theme_pubr ( ) +
theme ( legend.position = "right" ,
plot.title = element_text ( hjust = .5 ) )
} lme_plot <- map2 ( list ( lme1 ,lme2 ,lme3 ,lm ) ,list ( "随机截距+随机斜率" ,"随机截距+固定斜率" ,"固定截距+随机斜率" ,"固定截距+固定斜率" ) ,plot_lme ) lme_plot #> [[1]] #>
#> [[2]]#>
#> [[3]]#>
#> [[4]]
Show the code anova ( lme1 ,lme2 ,lme3 ) #> Data: df_long #> Models: #> lme2: Richness ~ 1 + NAP + (1 | Beach) #> lme3: Richness ~ 1 + NAP + (0 + NAP | Beach) #> lme1: Richness ~ 1 + NAP * Exposure + (1 + NAP | Beach) #> npar AIC BIC logLik deviance Chisq Df Pr(>Chisq) #> lme2 4 249.83 257.06 -120.92 241.83 #> lme3 4 261.95 269.18 -126.98 253.95 0.000 0 #> lme1 10 238.20 256.27 -109.10 218.20 35.754 6 3.077e-06 *** #> --- #> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 anova ( lme1 ,lme2 ,lme3 ,lme4 ,lm ) #> Data: df_long #> Models: #> lme4: Richness ~ 1 + (1 | Beach) #> lm: Richness ~ 1 + NAP #> lme2: Richness ~ 1 + NAP + (1 | Beach) #> lme3: Richness ~ 1 + NAP + (0 + NAP | Beach) #> lme1: Richness ~ 1 + NAP * Exposure + (1 + NAP | Beach) #> npar AIC BIC logLik deviance Chisq Df Pr(>Chisq) #> lme4 3 269.30 274.72 -131.65 263.30 #> lm 3 259.95 265.37 -126.98 253.95 9.350 0 #> lme2 4 249.83 257.06 -120.92 241.83 12.124 1 0.0004977 *** #> lme3 4 261.95 269.18 -126.98 253.95 0.000 0 #> lme1 10 238.20 256.27 -109.10 218.20 35.754 6 3.077e-06 *** #> --- #> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 # p小于0.05,说明全模型与简化后模型存在差异,最终采用lme1,AIC
模型诊断
sleepstudy
Show the code # 拟合线性混合模型 model <- lme1 # 1. 残差图 residuals <- resid ( model ) fitted <- fitted ( model ) ggplot ( data.frame ( fitted , residuals ) , aes ( fitted , residuals ) ) + geom_point ( ) +
geom_smooth ( method = "loess" , se = FALSE ) +
labs ( title = "Residuals vs Fitted" , x = "Fitted values" , y = "Residuals" ) Show the code
Show the code # 3. Cook's 距离 cooksd <- cooks.distance ( model ) plot ( cooksd , type = "h" , main = "Cook's Distance" ) Show the code # 4. 随机效应的分布 rand_dist <- ranef ( model ) qqnorm ( rand_dist $ Beach $ NAP ) qqline ( rand_dist $ Beach $ NAP )