11 非参数检验
11.1 秩
\[ F(Median)=P(X\le Median)=0.5 \]
11.2 二项分布B(n,0.5)
Show the code
library(ggplot2)
library(tibble)
library(dplyr)
library(readxl)
tibble(
x = -5:15,
y_binom = dbinom(x, size = 10,prob = 0.5),
) |>
ggplot()+
geom_col(aes(x=x,y=y_binom,color="binomal Distribution"),fill=NA)+
geom_function(mapping = aes(color="normal Distribution"),
fun = dnorm, args = list(mean = 5, sd = 1),
)+
scale_color_manual(values = c("normal Distribution" = "red",
"binomal Distribution" = "blue"))+
labs(color = "Distribution")
11.3 单样本 Wilcoxon Signed-Rank exact test
如果样本数据没有通过正态分布检验就要采用单样本wilcoxon符号秩检验进行计算。使用该检验需要满足的条件是样本值均匀地分布在均值两侧。
Show the code
shapiro.test(x)
#>
#> Shapiro-Wilk normality test
#>
#> data: x
#> W = 0.95237, p-value = 0.001192
wilcox.test(x, mu=7)
#>
#> Wilcoxon signed rank test with continuity correction
#>
#> data: x
#> V = 2518, p-value = 0.9822
#> alternative hypothesis: true location is not equal to 711.4 双样本
11.4.1 配对 Wilcoxon’s signed-rank test
\[ T_++T_-=\frac{n(n+1)}{2},n为非零配对差值的数量 \]
\[ T=min{(T_+,T_-)} \]
5 ≤ n ≤30,附表T0
n>16,正态近似法
Show the code
df <- tibble(
low=c(958.5,838.4,612.2,812.9,739.0,899.4,758.5,695.0,749.7,815.5),
high=c(958.5,866.5,788.9,815.2,783.2,910.9,760.8,870.8,862.3,799.9),
)
shapiro.test(df$high-df$low)
#>
#> Shapiro-Wilk normality test
#>
#> data: df$high - df$low
#> W = 0.79689, p-value = 0.01329
# 忽略 差异绝对值为“0”的数剔除;
wilcox.test(df$low[-1],df$high[-1],exact = T,paired = T)
#>
#> Wilcoxon signed rank exact test
#>
#> data: df$low[-1] and df$high[-1]
#> V = 4, p-value = 0.02734
#> alternative hypothesis: true location shift is not equal to 011.4.2 独立 Wilcoxon’s Rank-Sum 检验 (Mann-Whitney U 检验)
当两个样本不满足正态分布时,使用Wilcoxon秩和检验进行非参数检验
用于比较两个独立样本的中位数是否相等。
\[ Wilxoxon秩和\ T=min\{T_1,T_2\} \]
Show the code
MVR = c(38, 29, 35, 33, 38, 41, 31)
MVP = c(32, 43, 44, 81, 35, 46, 37, 45, 44)
shapiro.test(c(MVR,MVP))
#>
#> Shapiro-Wilk normality test
#>
#> data: c(MVR, MVP)
#> W = 0.70443, p-value = 0.0001889
combined_data <- c(MVR, MVP)
ranked_data <- rank(combined_data)
ranked_data
#> [1] 8.5 1.0 5.5 4.0 8.5 10.0 2.0 3.0 11.0 12.5 16.0 5.5 15.0 7.0 14.0
#> [16] 12.5
MVR_ranks <- ranked_data[1:length(MVR)]
MVP_ranks <- ranked_data[(length(MVR)+1):length(combined_data)]
T1 <- sum(MVR_ranks)
T2 <- sum(MVP_ranks)
T1-length(MVR)*(length(MVR)+1)/2
#> [1] 11.5
wilcox.test(MVR,MVP,exact = F,correct = F)
#>
#> Wilcoxon rank sum test
#>
#> data: MVR and MVP
#> W = 11.5, p-value = 0.03386
#> alternative hypothesis: true location shift is not equal to 011.4.2.1 W统计量
n1<10,n2-n1<10,附录
n1>10,n2>10,正态近似法
Show the code
x <- c(17, 12, 13, 16, 9, 19, 21, 12, 18, 17)
y <- c(10, 6, 15, 9, 8, 11, 8, 16, 13, 7, 5, 14)
wilcox.test(x, y, correct = F)
#>
#> Wilcoxon rank sum test
#>
#> data: x and y
#> W = 101.5, p-value = 0.006124
#> alternative hypothesis: true location shift is not equal to 0
rank(c(x,y))[1:10] |> sum()
#> [1] 156.5
rank(c(x,y))[11:22] |> sum()
#> [1] 96.5
156.5-10*11/2
#> [1] 101.5
a <- wilcox.test(x,y,correct=FALSE)
str(a)
#> List of 7
#> $ statistic : Named num 102
#> ..- attr(*, "names")= chr "W"
#> $ parameter : NULL
#> $ p.value : num 0.00612
#> $ null.value : Named num 0
#> ..- attr(*, "names")= chr "location shift"
#> $ alternative: chr "two.sided"
#> $ method : chr "Wilcoxon rank sum test"
#> $ data.name : chr "x and y"
#> - attr(*, "class")= chr "htest"
n1 <- length(x)
a$statistic <- a$statistic + n1*(n1+1)/2
names(a$statistic) <- "T.W"
a
#>
#> Wilcoxon rank sum test
#>
#> data: x and y
#> T.W = 156.5, p-value = 0.006124
#> alternative hypothesis: true location shift is not equal to 011.4.2.2 曼-惠特尼 U 统计量
\[ 曼-惠特尼U统计量= 威尔科克森W(较小秩和)-\frac{n_{T_{min}}(n_{T_{min}}+1)}{2} \]
11.4.2.3 Z 统计量 coin::wilcox_test()
Show the code
library(coin)
df <- read_excel("data/coin-wilcox_test.xlsx") |>
mutate(group=as.factor(group))
df
#> # A tibble: 240 × 2
#> HADS得分 group
#> <dbl> <fct>
#> 1 14 1
#> 2 14 1
#> 3 7 1
#> 4 22 1
#> 5 7 1
#> 6 12 1
#> 7 15 1
#> 8 17 1
#> 9 8 1
#> 10 14 1
#> # ℹ 230 more rows
rank <- rank(df$HADS得分)
g1rankSum <- sum(rank[1:120])
g2rankSum <- sum(rank[121:240])
SPSS_威尔科克森W <- min(g1rankSum,g2rankSum)
SPSS_威尔科克森W
#> [1] 13965.5
SPSS_曼惠特尼U <- SPSS_威尔科克森W -120*121/2
SPSS_曼惠特尼U
#> [1] 6705.5
wilcox.test(HADS得分 ~ group,data=df,correct=F)
#>
#> Wilcoxon rank sum test
#>
#> data: HADS得分 by group
#> W = 7694.5, p-value = 0.3572
#> alternative hypothesis: true location shift is not equal to 0
# SPSS z统计量
coin::wilcox_test( HADS得分 ~ group,data=df, distribution = "asymptotic") # exact asymptotic approximate
#>
#> Asymptotic Wilcoxon-Mann-Whitney Test
#>
#> data: HADS得分 by group (1, 2)
#> Z = 0.92062, p-value = 0.3572
#> alternative hypothesis: true mu is not equal to 0
coin::wilcox_test( HADS得分 ~ group,data=df, distribution = "approximate")
#>
#> Approximative Wilcoxon-Mann-Whitney Test
#>
#> data: HADS得分 by group (1, 2)
#> Z = 0.92062, p-value = 0.3589
#> alternative hypothesis: true mu is not equal to 0SPSS 用较小秩和减去对应 n(n+1)/2
R有时用较小秩和减去对应 n(n+1)/2,有时用较大秩和减去对应 n(n+1)/2
Show the code
wilcox.test(HADS得分 ~ group,data=df,exact = F,correct = F)
#>
#> Wilcoxon rank sum test
#>
#> data: HADS得分 by group
#> W = 7694.5, p-value = 0.3572
#> alternative hypothesis: true location shift is not equal to 0
# SPSS_曼惠特尼U
g2rankSum-120*121/2
#> [1] 6705.5
# R中W
g1rankSum-120*121/2
#> [1] 7694.5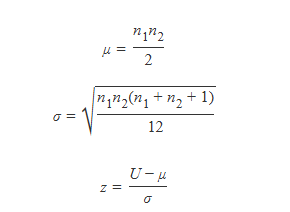
n1>20
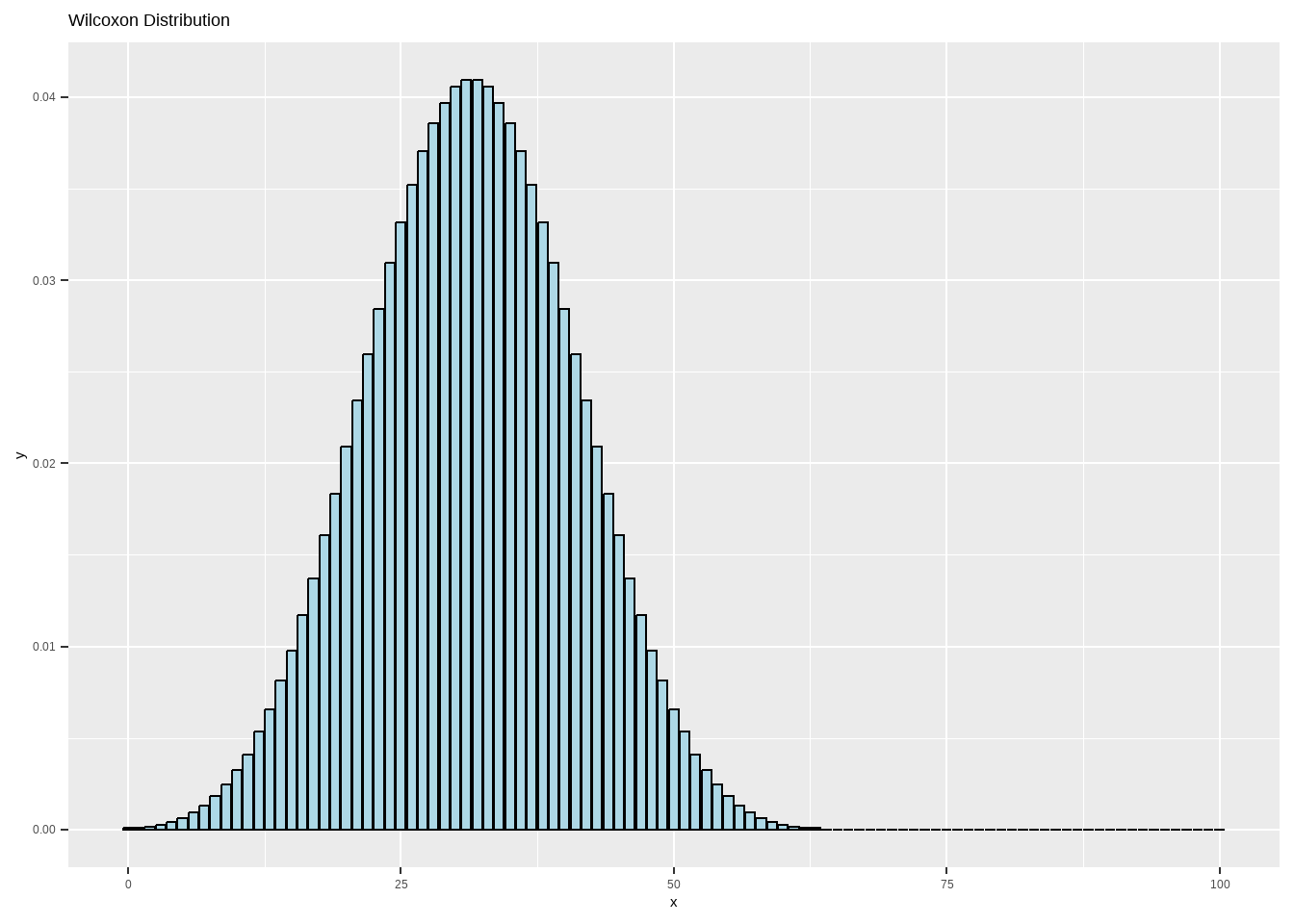
11.4.3 Wilcoxon Distribution
11.5 多样本
11.5.1 独立 Kruskal-Wallis 检验
用于比较三个或更多独立样本的中位数是否相等。
假设:
随机,独立
每个样本至少5个观测
能够计算秩次
Show the code
kruskal.test(weight~group,data = PlantGrowth)
#>
#> Kruskal-Wallis rank sum test
#>
#> data: weight by group
#> Kruskal-Wallis chi-squared = 7.9882, df = 2, p-value = 0.0184211.5.1.1 事后多重比较
Show the code
pairwise.wilcox.test(PlantGrowth$weight,PlantGrowth$group,p.adjust.method = "fdr",exact=F)
#>
#> Pairwise comparisons using Wilcoxon rank sum test with continuity correction
#>
#> data: PlantGrowth$weight and PlantGrowth$group
#>
#> ctrl trt1
#> trt1 0.199 -
#> trt2 0.096 0.034
#>
#> P value adjustment method: fdr11.6 相关 Friedman 检验
用于比较三个或更多相关样本的中位数是否相等。
Show the code
# 假设有三个相关样本 x, y, z
x <- c(14, 17, 20, 23, 25)
y <- c(15, 18, 21, 24, 26)
z <- c(16, 19, 22, 25, 27)
# 将样本合并成一个数据框,并指定组别和受试者
data <- data.frame(
value = c(x, y, z),
group = factor(rep(c("x", "y", "z"), each = 5)),
subject = factor(rep(1:5, 3))
)
# 使用 friedman.test() 函数进行检验
result <- friedman.test(value ~ group | subject, data = data)
# 输出检验结果
print(result)
#>
#> Friedman rank sum test
#>
#> data: value and group and subject
#> Friedman chi-squared = 10, df = 2, p-value = 0.00673811.7 Kendall’s Tau 检验
用途:用于检验两个变量之间的相关性。
Show the code
# 假设有两个变量 x 和 y
x <- c(14, 17, 20, 23, 25)
y <- c(15, 18, 21, 24, 26)
# 使用 cor.test() 函数进行 Kendall's Tau 检验
result <- cor.test(x, y, method = "kendall")
# 输出检验结果
print(result)
#>
#> Kendall's rank correlation tau
#>
#> data: x and y
#> T = 10, p-value = 0.01667
#> alternative hypothesis: true tau is not equal to 0
#> sample estimates:
#> tau
#> 111.8 Spearman’s Rank Correlation 检验
用途:用于检验两个变量之间的相关性,适用于数据非线性关系。
Show the code
# 假设有两个变量 x 和 y
x <- c(14, 17, 20, 23, 25)
y <- c(15, 18, 21, 24, 26)
# 使用 cor.test() 函数进行 Spearman's Rank Correlation 检验
result <- cor.test(x, y, method = "spearman")
# 输出检验结果
print(result)
#>
#> Spearman's rank correlation rho
#>
#> data: x and y
#> S = 4.4409e-15, p-value = 0.01667
#> alternative hypothesis: true rho is not equal to 0
#> sample estimates:
#> rho
#> 1