线性混合效应模型 (Linear Mixed-effect Model,LMM)用于分析具有固定效应和随机效应的数据结构。 它能够处理数据中的个体异质性和组内相关性。如果数据满足正态分布但有组内相关性 ,使用线性混合效应模型。
线性混合模型的一般表达形式:
\[ Y = \mathbf{X} \beta + \mathbf{Z} \gamma + \epsilon \]
其中:
固定效应是感兴趣的自变量,表示所有个体都相同的效应
随机效应:随机截距表示不同个体的基线差异,随机斜率表示不同个体对自变量的响应差异。
模型假设Cov(γ,X)=0
因变量 Y 为连续型数值变量
线性
残差的正态性 \(\epsilon\sim N(0,\sigma^2 I)\)
方差齐性 \(Var(ε_i )=σ^2\)
随机效应假设
随机效应与「所有固定效应自变量」之间相互独立 \(Cov(γ,X)=0\)
随机效应 ( γ ) 服从均值为 0 的多元正态分布 \(γ∼N(0,Σ_γ )\)
随机效应 ( γ ) 与残差 ( ε ) 之间相互独立 \(Cov(γ,ε)=0\)
随机效应的方差-协方差矩阵\(Σ_γ\) ,残差的方差矩阵 \(σ^2I\) 的结构相关性特征:
对于连续数据,mixed model repeated measures(MMRM)
对于分类数据或计数数据,generalized linear mixed-effects model(GLMM)
Mixed Effects Models and Extensions in Ecology with R 第5章
lme4::lmer()
lmer()
\[
lmer (data,formual= DV \sim Fixed\_Factor + \\ (Random\_intercept + Random\_slope | Random\_Factor)+\\(Random\_intercept + Random\_slope | Random\_Factor)+...
\\)
\]
截距中,1表示随机截距,0表示固定截距,默认截距为1。
随机截距+随机斜率
y~x+( 1+x | id )
y~x+( x | id )
随机截距+固定斜率
y~x+( 1+1 | id )
y~x+( 1 | id )
固定截距+随机斜率
y~x+( 0+x | id )
NA
线性模型:固定截距+固定斜率
y~x
NA
Show the code library ( readr ) library ( dplyr ) rikz <- read_tsv ( "data/AED/RIKZ.txt" ) rikz <- rikz |> mutate (
Beach = factor ( Beach ) ,
Exposure = factor ( Exposure )
)
head ( rikz ) #> # A tibble: 6 × 5 #> Sample Richness Exposure NAP Beach #> <dbl> <dbl> <fct> <dbl> <fct> #> 1 1 11 10 0.045 1 #> 2 2 10 10 -1.04 1 #> 3 3 13 10 -1.34 1 #> 4 4 11 10 0.616 1 #> 5 5 10 10 -0.684 1 #> 6 6 8 8 1.19 2
随机截距+随机斜率
\[
\eta_{(nrow \times 1)} = \mathbf{X}_{nrow \times 1} \beta_{1 \times 1} + \mathbf{Z}_{nrow \times 2n_{subjects}} \mathbf{\gamma}_{2n_{subjects} \times 1} + \epsilon_i
\]
Z有两倍于受试者数量的列,每个受试者的随机截距和随机斜率
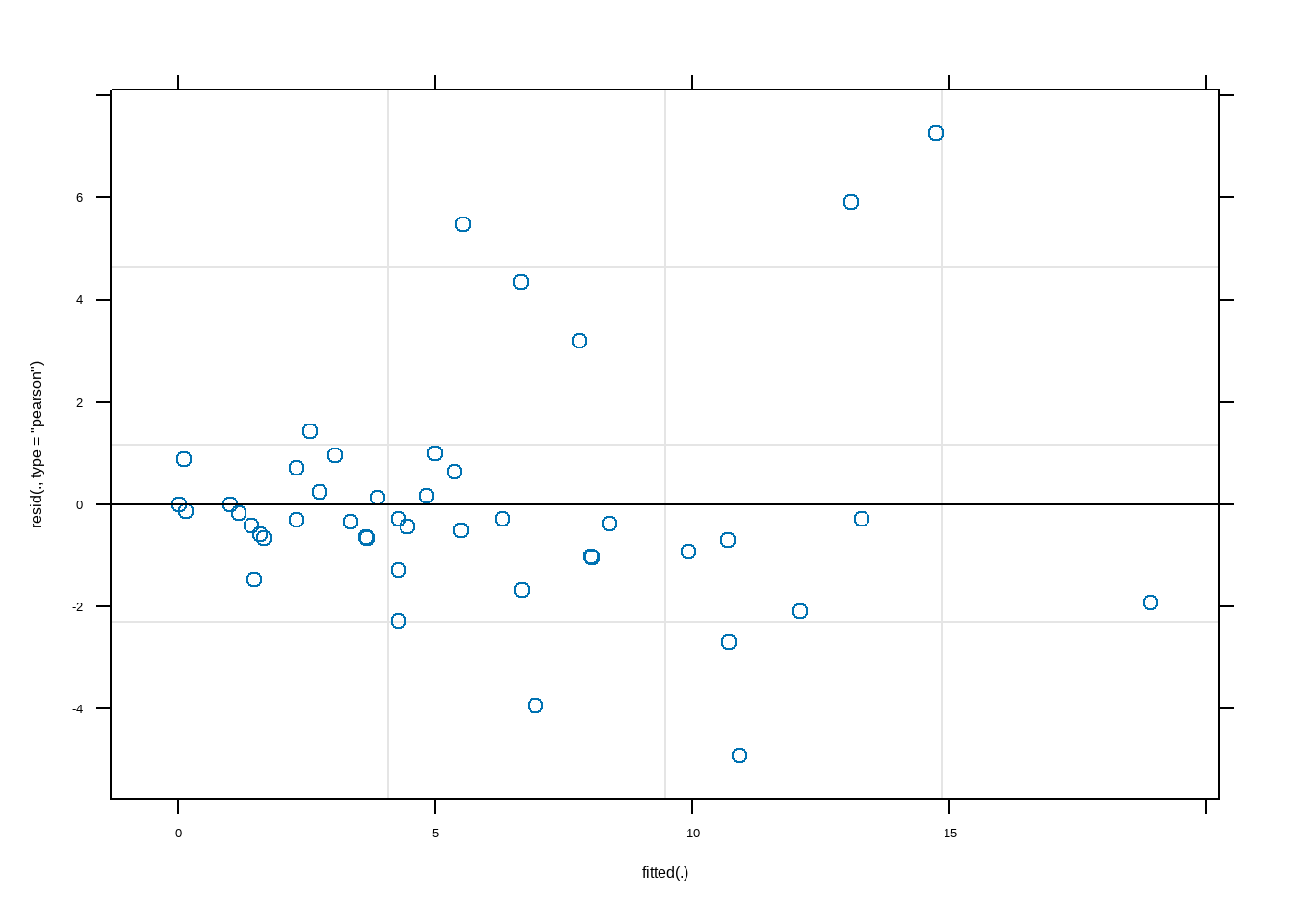
Show the code library ( lmerTest ) conflicted :: conflicts_prefer ( lmerTest :: lmer ) lme1 <- lmer ( Richness ~ 1 + NAP + Exposure + NAP : Exposure + ( 1 + NAP | Beach ) ,data = rikz ) summary ( lme1 ) #> Linear mixed model fit by REML. t-tests use Satterthwaite's method [ #> lmerModLmerTest] #> Formula: Richness ~ 1 + NAP + Exposure + NAP:Exposure + (1 + NAP | Beach) #> Data: rikz #> #> REML criterion at convergence: 207.2 #> #> Scaled residuals: #> Min 1Q Median 3Q Max #> -1.92384 -0.36066 -0.13343 0.09819 2.84228 #> #> Random effects: #> Groups Name Variance Std.Dev. Corr #> Beach (Intercept) 3.758 1.938 #> NAP 2.837 1.684 -1.00 #> Residual 6.535 2.556 #> Number of obs: 45, groups: Beach, 9 #> #> Fixed effects: #> Estimate Std. Error df t value Pr(>|t|) #> (Intercept) 13.3457 2.2784 5.0864 5.858 0.00194 ** #> NAP -4.1753 2.1243 5.7080 -1.965 0.09942 . #> Exposure10 -5.3273 2.5556 5.1415 -2.085 0.09000 . #> Exposure11 -9.7660 2.5653 5.1903 -3.807 0.01169 * #> NAP:Exposure10 0.1646 2.3621 5.5602 0.070 0.94688 #> NAP:Exposure11 2.7273 2.3715 5.6329 1.150 0.29661 #> --- #> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 #> #> Correlation of Fixed Effects: #> (Intr) NAP Exps10 Exps11 NAP:E10 #> NAP -0.770 #> Exposure10 -0.892 0.686 #> Exposure11 -0.888 0.683 0.792 #> NAP:Expsr10 0.692 -0.899 -0.775 -0.615 #> NAP:Expsr11 0.689 -0.896 -0.615 -0.779 0.806 #> optimizer (nloptwrap) convergence code: 0 (OK) #> boundary (singular) fit: see help('isSingular') plot ( lme1 ) Show the code AIC ( lme1 ) #> [1] 227.1558 BIC ( lme1 ) #> [1] 245.2224 logLik ( lme1 ) #> 'log Lik.' -103.5779 (df=10) # library(broom.mixed) # tidy(lme1) library ( nlme ) lme ( Richness ~ 1 + NAP * Exposure , random = ~ 1 + NAP | Beach ,
data = rikz ,
control = lmeControl ( opt = "optim" ,
msMaxIter = 1000 ,
msMaxEval = 5000 )
) |>
summary ( )
#> Linear mixed-effects model fit by REML #> Data: rikz #> AIC BIC logLik #> 227.2046 243.8402 -103.6023 #> #> Random effects: #> Formula: ~1 + NAP | Beach #> Structure: General positive-definite, Log-Cholesky parametrization #> StdDev Corr #> (Intercept) 1.939709 (Intr) #> NAP 1.689580 -0.996 #> Residual 2.554676 #> #> Fixed effects: Richness ~ 1 + NAP * Exposure #> Value Std.Error DF t-value p-value #> (Intercept) 13.345694 2.278989 33 5.855970 0.0000 #> NAP -4.175271 2.128084 33 -1.961986 0.0582 #> Exposure10 -5.323695 2.556406 6 -2.082492 0.0825 #> Exposure11 -9.765613 2.566035 6 -3.805720 0.0089 #> NAP:Exposure10 0.167146 2.366467 33 0.070631 0.9441 #> NAP:Exposure11 2.726865 2.375786 33 1.147774 0.2593 #> Correlation: #> (Intr) NAP Exps10 Exps11 NAP:E10 #> NAP -0.767 #> Exposure10 -0.891 0.684 #> Exposure11 -0.888 0.682 0.792 #> NAP:Exposure10 0.690 -0.899 -0.772 -0.613 #> NAP:Exposure11 0.687 -0.896 -0.613 -0.777 0.806 #> #> Standardized Within-Group Residuals: #> Min Q1 Med Q3 Max #> -1.9251786 -0.3608959 -0.1327685 0.0992999 2.8420571 #> #> Number of Observations: 45 #> Number of Groups: 9
这个模型的公式可以分解为:
REML 限制性最大似然准则 207.2,评估模型的拟合优度
Scaled residuals
随机效应部分 :Beach 作为随机效应的分组变量,包含随机截距和随机斜率(NAP),相关性为 -1,说明随机截距和随机斜率之间呈现完全负相关关系。
固定效应部分 :Richness 的预测由截距、NAP(数值变量)和 Exposure(分类变量,包含 Exposure10 和 Exposure11)及其交互项构成。
固定效应的相关性矩阵
Show the code # 标准化模型残差分布 quantile ( residuals ( lme1 ,type= "pearson" ,scaled= T ) ) #> 0% 25% 50% 75% 100% #> -1.9238372 -0.3606612 -0.1334336 0.0981939 2.8422803 # 查看固定效应和显著性检验 fixef ( lme1 ) #> (Intercept) NAP Exposure10 Exposure11 NAP:Exposure10 #> 13.3456944 -4.1752712 -5.3272654 -9.7660482 0.1646123 #> NAP:Exposure11 #> 2.7273068 anova ( lme1 ) #> Type III Analysis of Variance Table with Satterthwaite's method #> Sum Sq Mean Sq NumDF DenDF F value Pr(>F) #> NAP 90.646 90.646 1 5.5230 13.8711 0.01139 * #> Exposure 109.852 54.926 2 5.2975 8.4051 0.02273 * #> NAP:Exposure 22.308 11.154 2 5.3880 1.7068 0.26657 #> --- #> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 # 随机效应:随机截距和随机斜率 # 每个组的随机截距表示该组的平均值与模型整体平均值(截距)的差异 ranef ( lme1 ) #> $Beach #> (Intercept) NAP #> 1 -4.274356e-02 3.713708e-02 #> 2 -2.709479e-14 2.354089e-14 #> 3 3.819348e-03 -3.318381e-03 #> 4 -2.741786e-01 2.382158e-01 #> 5 3.269522e+00 -2.840673e+00 #> 6 2.736093e-01 -2.377212e-01 #> 7 -3.250033e-03 2.823741e-03 #> 8 -2.148947e+00 1.867079e+00 #> 9 -1.077831e+00 9.364564e-01 #> #> with conditional variances for "Beach" # 随机效应的显著性检验 lmerTest :: ranova ( lme1 ) #> ANOVA-like table for random-effects: Single term deletions #> #> Model: #> Richness ~ NAP + Exposure + (1 + NAP | Beach) + NAP:Exposure #> npar logLik AIC LRT Df Pr(>Chisq) #> <none> 10 -103.58 227.16 #> NAP in (1 + NAP | Beach) 8 -105.67 227.35 4.1935 2 0.1229 cor ( ranef ( lme1 ) $ Beach ) #> (Intercept) NAP #> (Intercept) 1 -1 #> NAP -1 1
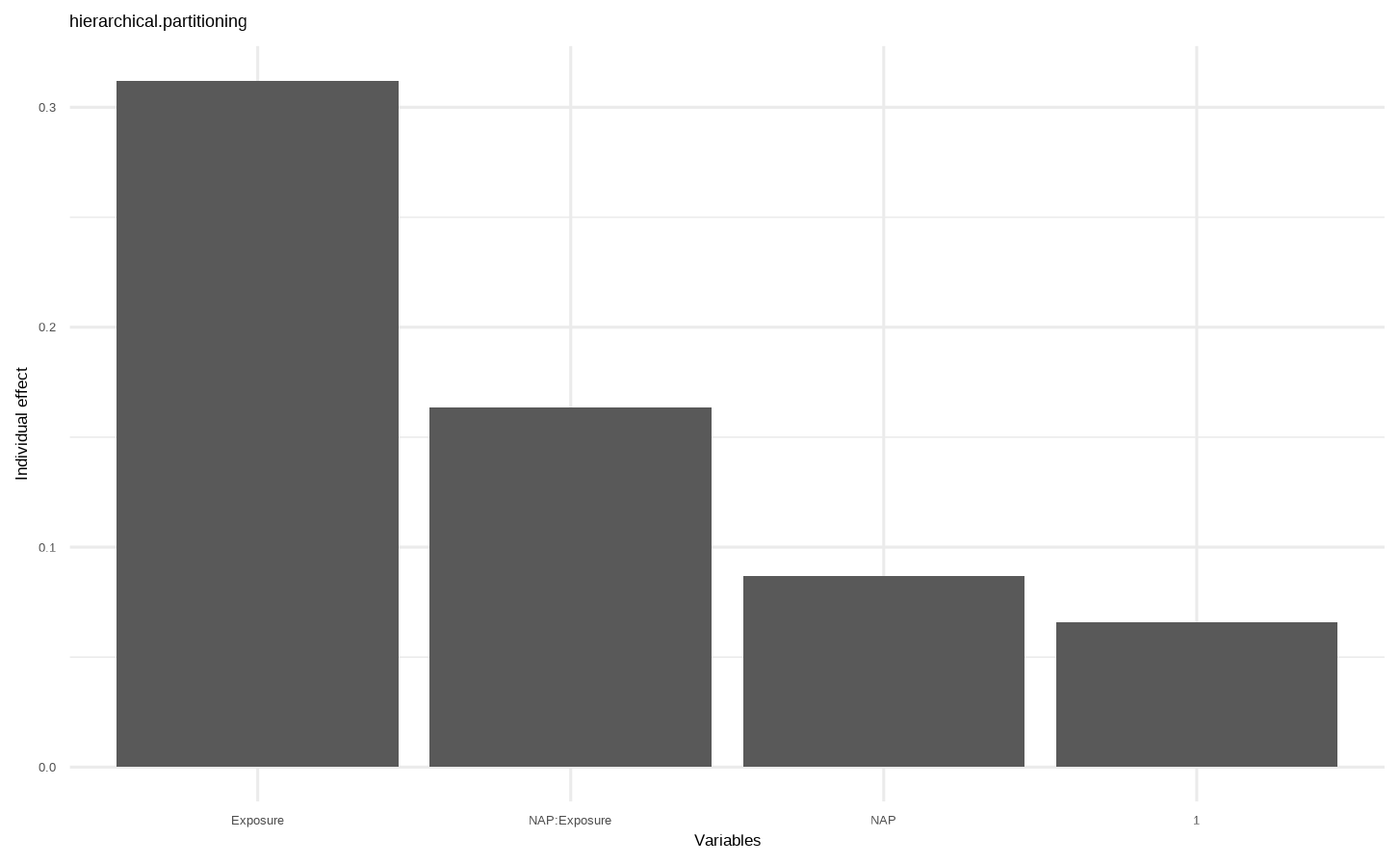
Show the code e <- glmm.hp :: glmm.hp ( lme1 ) save ( e ,file = "data/glmm.hp.Rdata" ) # $r.squaredGLMM # R2m R2c # [1,] 0.6279508 0.7811086 # # $hierarchical.partitioning # Unique Average.share Individual I.perc(%) # 1 0.0000 0.0657 0.0657 10.46 # NAP 0.0000 0.0867 0.0867 13.81 # Exposure 0.2987 0.0135 0.3122 49.71 # NAP:Exposure 0.0884 0.0750 0.1634 26.02 # # $variables # [1] "1" "NAP" "Exposure" "NAP:Exposure" # # $type # [1] "hierarchical.partitioning" # # attr(,"class") # [1] "glmmhp"
Show the code load ( "data/glmm.hp.Rdata" ) e #> $r.squaredGLMM #> R2m R2c #> [1,] 0.6279508 0.7811086 #> #> $hierarchical.partitioning #> Unique Average.share Individual I.perc(%) #> 1 0.0000 0.0657 0.0657 10.46 #> NAP 0.0000 0.0867 0.0867 13.81 #> Exposure 0.2987 0.0135 0.3122 49.71 #> NAP:Exposure 0.0884 0.0750 0.1634 26.02 #> #> $variables #> [1] "1" "NAP" "Exposure" "NAP:Exposure" #> #> $type #> [1] "hierarchical.partitioning" #> #> attr(,"class") #> [1] "glmmhp"
$r.squaredGLMM
$hierarchical.partitioning
“Unique” 列 :展示了各变量独自解释的方差比例
“Average.share” 列 :变量与其他变量所有组合的方差解释比例
“Individual” 列 :“Unique”和“Average.share”的和,即变量的总贡献
“I.perc (%)” 列 :每个变量的相对重要性程度。
$variables
简单列出了模型中涉及的变量名称,这里有 "1"(代表整体截距)、"NAP"、"Exposure" 以及它们的交互项 "NAP:Exposure",明确了模型在分析中考虑的因素构成。
$type
指出了当前分析结果呈现的类型是 "hierarchical.partitioning",说明后续如果进一步拓展分析或者对比不同模型等操作时,可以清楚知道这部分结果对应的分析类别。
随机截距+固定斜率
\[
\eta_{(nrow \ \times 1)}=\mathbf{X_{nrow×1}}\beta_{1\times1} +Z_{nrow\times n_{subjects}} \mathbf{\gamma}_{n_{subjects}\times 1}+\epsilon_i
\]
Z有一倍于受试者数量的列,每个受试者的随机截距。
Show the code lme2 <- lmer ( Richness ~ 1 + NAP * Exposure + ( 1 + 1 | Beach ) ,data = rikz ) summary ( lme2 ) #> Linear mixed model fit by REML. t-tests use Satterthwaite's method [ #> lmerModLmerTest] #> Formula: Richness ~ 1 + NAP * Exposure + (1 + 1 | Beach) #> Data: rikz #> #> REML criterion at convergence: 211.3 #> #> Scaled residuals: #> Min 1Q Median 3Q Max #> -1.5653 -0.4387 -0.1165 0.1783 4.1098 #> #> Random effects: #> Groups Name Variance Std.Dev. #> Beach (Intercept) 0.2641 0.5139 #> Residual 8.8316 2.9718 #> Number of obs: 45, groups: Beach, 9 #> #> Fixed effects: #> Estimate Std. Error df t value Pr(>|t|) #> (Intercept) 13.3457 1.4836 6.5050 8.996 6.63e-05 *** #> NAP -4.1753 1.5051 33.0857 -2.774 0.009030 ** #> Exposure10 -5.5450 1.6577 6.4545 -3.345 0.013909 * #> Exposure11 -9.7306 1.6705 6.6476 -5.825 0.000781 *** #> NAP:Exposure10 0.6717 1.6439 34.9341 0.409 0.685306 #> NAP:Exposure11 2.6888 1.6567 34.2669 1.623 0.113766 #> --- #> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 #> #> Correlation of Fixed Effects: #> (Intr) NAP Exps10 Exps11 NAP:E10 #> NAP -0.278 #> Exposure10 -0.895 0.249 #> Exposure11 -0.888 0.247 0.795 #> NAP:Expsr10 0.255 -0.916 -0.276 -0.226 #> NAP:Expsr11 0.253 -0.908 -0.226 -0.296 0.832 AIC ( lme2 ) #> [1] 227.3493 BIC ( lme2 ) #> [1] 241.8026 logLik ( lme2 ) #> 'log Lik.' -105.6747 (df=8) 2 * ( 1 - pt ( 6.007 ,35 ,lower.tail = T ) ) #> [1] 7.558855e-07 lme ( Richness ~ 1 + NAP * Exposure ,random= ~ 1 + 1 | Beach ,data = rikz ) |> summary ( )
#> Linear mixed-effects model fit by REML #> Data: rikz #> AIC BIC logLik #> 227.3493 240.6578 -105.6747 #> #> Random effects: #> Formula: ~1 + 1 | Beach #> (Intercept) Residual #> StdDev: 0.5138683 2.971793 #> #> Fixed effects: Richness ~ 1 + NAP * Exposure #> Value Std.Error DF t-value p-value #> (Intercept) 13.345694 1.483557 33 8.995739 0.0000 #> NAP -4.175271 1.505110 33 -2.774063 0.0090 #> Exposure10 -5.544983 1.657659 6 -3.345069 0.0155 #> Exposure11 -9.730595 1.670518 6 -5.824898 0.0011 #> NAP:Exposure10 0.671731 1.643864 33 0.408629 0.6855 #> NAP:Exposure11 2.688806 1.656743 33 1.622947 0.1141 #> Correlation: #> (Intr) NAP Exps10 Exps11 NAP:E10 #> NAP -0.278 #> Exposure10 -0.895 0.249 #> Exposure11 -0.888 0.247 0.795 #> NAP:Exposure10 0.255 -0.916 -0.276 -0.226 #> NAP:Exposure11 0.253 -0.908 -0.226 -0.296 0.832 #> #> Standardized Within-Group Residuals: #> Min Q1 Med Q3 Max #> -1.5652904 -0.4386841 -0.1164805 0.1783113 4.1098230 #> #> Number of Observations: 45 #> Number of Groups: 9
固定截距+随机斜率
\[
\eta_{(nrow \times 1)} = \mathbf{X}_{nrow \times 1} \beta_{1 \times 1} + \mathbf{Z}_{nrow \times n_{subjects}} \mathbf{\gamma}_{n_{subjects} \times 1} + \epsilon_i
\]
Z 有一倍于受试者数量的列,每个受试者的随机斜率。
Show the code lme3 <- lmer ( Richness ~ 1 + NAP * Exposure + ( 0 + NAP | Beach ) ,data = rikz ) summary ( lme3 ) #> Linear mixed model fit by REML. t-tests use Satterthwaite's method [ #> lmerModLmerTest] #> Formula: Richness ~ 1 + NAP * Exposure + (0 + NAP | Beach) #> Data: rikz #> #> REML criterion at convergence: 211.4 #> #> Scaled residuals: #> Min 1Q Median 3Q Max #> -1.6048 -0.4448 -0.1226 0.2322 4.1284 #> #> Random effects: #> Groups Name Variance Std.Dev. #> Beach NAP 0.000 0.000 #> Residual 9.024 3.004 #> Number of obs: 45, groups: Beach, 9 #> #> Fixed effects: #> Estimate Std. Error df t value Pr(>|t|) #> (Intercept) 13.3457 1.4068 39.0000 9.486 1.11e-11 *** #> NAP -4.1753 1.5214 39.0000 -2.744 0.00912 ** #> Exposure10 -5.5318 1.5715 39.0000 -3.520 0.00111 ** #> Exposure11 -9.7284 1.5852 39.0000 -6.137 3.34e-07 *** #> NAP:Exposure10 0.6278 1.6583 39.0000 0.379 0.70703 #> NAP:Exposure11 2.6835 1.6725 39.0000 1.604 0.11668 #> --- #> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 #> #> Correlation of Fixed Effects: #> (Intr) NAP Exps10 Exps11 NAP:E10 #> NAP -0.297 #> Exposure10 -0.895 0.266 #> Exposure11 -0.887 0.263 0.794 #> NAP:Expsr10 0.272 -0.917 -0.294 -0.242 #> NAP:Expsr11 0.270 -0.910 -0.242 -0.315 0.835 #> optimizer (nloptwrap) convergence code: 0 (OK) #> boundary (singular) fit: see help('isSingular') lme ( Richness ~ 1 + NAP * Exposure ,random= ~ 0 + NAP | Beach ,data = rikz ) |> summary ( )
#> Linear mixed-effects model fit by REML #> Data: rikz #> AIC BIC logLik #> 227.3962 240.7047 -105.6981 #> #> Random effects: #> Formula: ~0 + NAP | Beach #> NAP Residual #> StdDev: 0.0001223769 3.004042 #> #> Fixed effects: Richness ~ 1 + NAP * Exposure #> Value Std.Error DF t-value p-value #> (Intercept) 13.345694 1.406821 33 9.486418 0.0000 #> NAP -4.175271 1.521443 33 -2.744283 0.0097 #> Exposure10 -5.531822 1.571461 6 -3.520178 0.0125 #> Exposure11 -9.728394 1.585241 6 -6.136854 0.0009 #> NAP:Exposure10 0.627847 1.658269 33 0.378616 0.7074 #> NAP:Exposure11 2.683487 1.672531 33 1.604447 0.1181 #> Correlation: #> (Intr) NAP Exps10 Exps11 NAP:E10 #> NAP -0.297 #> Exposure10 -0.895 0.266 #> Exposure11 -0.887 0.263 0.794 #> NAP:Exposure10 0.272 -0.917 -0.294 -0.242 #> NAP:Exposure11 0.270 -0.910 -0.242 -0.315 0.835 #> #> Standardized Within-Group Residuals: #> Min Q1 Med Q3 Max #> -1.6048271 -0.4448474 -0.1225590 0.2321686 4.1283628 #> #> Number of Observations: 45 #> Number of Groups: 9
随机效应模型
Show the code lme4 <- lmer ( Richness ~ 1 + ( 1 + NAP | Beach ) ,data = rikz ) summary ( lme4 ) #> Linear mixed model fit by REML. t-tests use Satterthwaite's method [ #> lmerModLmerTest] #> Formula: Richness ~ 1 + (1 + NAP | Beach) #> Data: rikz #> #> REML criterion at convergence: 242.9 #> #> Scaled residuals: #> Min 1Q Median 3Q Max #> -2.1607 -0.4415 -0.1999 0.2065 2.6133 #> #> Random effects: #> Groups Name Variance Std.Dev. Corr #> Beach (Intercept) 28.173 5.308 #> NAP 11.292 3.360 -0.95 #> Residual 6.697 2.588 #> Number of obs: 45, groups: Beach, 9 #> #> Fixed effects: #> Estimate Std. Error df t value Pr(>|t|) #> (Intercept) 2.8023 0.8218 3.7478 3.41 0.0299 * #> --- #> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ranef ( lme4 ) #> $Beach #> (Intercept) NAP #> 1 6.0112287 -2.8605659 #> 2 9.7420133 -5.0379262 #> 3 0.8792316 -0.9838634 #> 4 0.3721784 -0.7548668 #> 5 9.1546337 -6.7668418 #> 6 1.5719076 -1.1861532 #> 7 0.4484228 -0.7999673 #> 8 2.1690889 -1.6492031 #> 9 3.3610981 -2.4639704 #> #> with conditional variances for "Beach" lme ( Richness ~ 1 ,random= ~ 1 + NAP | Beach ,data = rikz ) |> summary ( )
#> Linear mixed-effects model fit by REML #> Data: rikz #> AIC BIC logLik #> 252.935 261.8559 -121.4675 #> #> Random effects: #> Formula: ~1 + NAP | Beach #> Structure: General positive-definite, Log-Cholesky parametrization #> StdDev Corr #> (Intercept) 5.307778 (Intr) #> NAP 3.360427 -0.948 #> Residual 2.587870 #> #> Fixed effects: Richness ~ 1 #> Value Std.Error DF t-value p-value #> (Intercept) 2.802324 0.8217849 36 3.410046 0.0016 #> #> Standardized Within-Group Residuals: #> Min Q1 Med Q3 Max #> -2.1606764 -0.4415215 -0.1998751 0.2064870 2.6133376 #> #> Number of Observations: 45 #> Number of Groups: 9
线性模型:固定截距+ 固定斜率
Show the code lm <- lm ( Richness ~ 1 + NAP * Exposure ,data = rikz ) lm #> #> Call: #> lm(formula = Richness ~ 1 + NAP * Exposure, data = rikz) #> #> Coefficients: #> (Intercept) NAP Exposure10 Exposure11 NAP:Exposure10 #> 13.3457 -4.1753 -5.5318 -9.7284 0.6278 #> NAP:Exposure11 #> 2.6835
模型选择
限制最大似然法REML
赤池信息准则(AIC)
\[ kIC=−2log(\mathcal{L})+2k\]
其中:
\(\mathcal{L}\) 是似然函数。
\(k\) 是模型参数的数量。
贝叶斯信息准则(BIC)
\[ BIC=−2log(\mathcal{L})+klog(n) \]
其中:
Show the code plot_lme <- function ( model , title ) { ggplot ( rikz , aes ( NAP , Richness , group = Beach , color = Beach ) ) +
geom_point ( ) +
geom_line (
data = bind_cols ( rikz , .pred = predict ( model , rikz ) ) ,
mapping = aes ( y = .pred ) ,
linewidth = 1
) +
labs ( title = title ) +
scale_x_continuous ( expand = ( mult = c ( 0 ,.1 ) ) ) +
scale_y_continuous ( expand = ( mult = c ( 0 ,.1 ) ) ) +
ggsci :: scale_color_jco ( ) +
ggpubr :: theme_pubr ( ) +
theme ( legend.position = "right" ,
plot.title = element_text ( hjust = .5 ) )
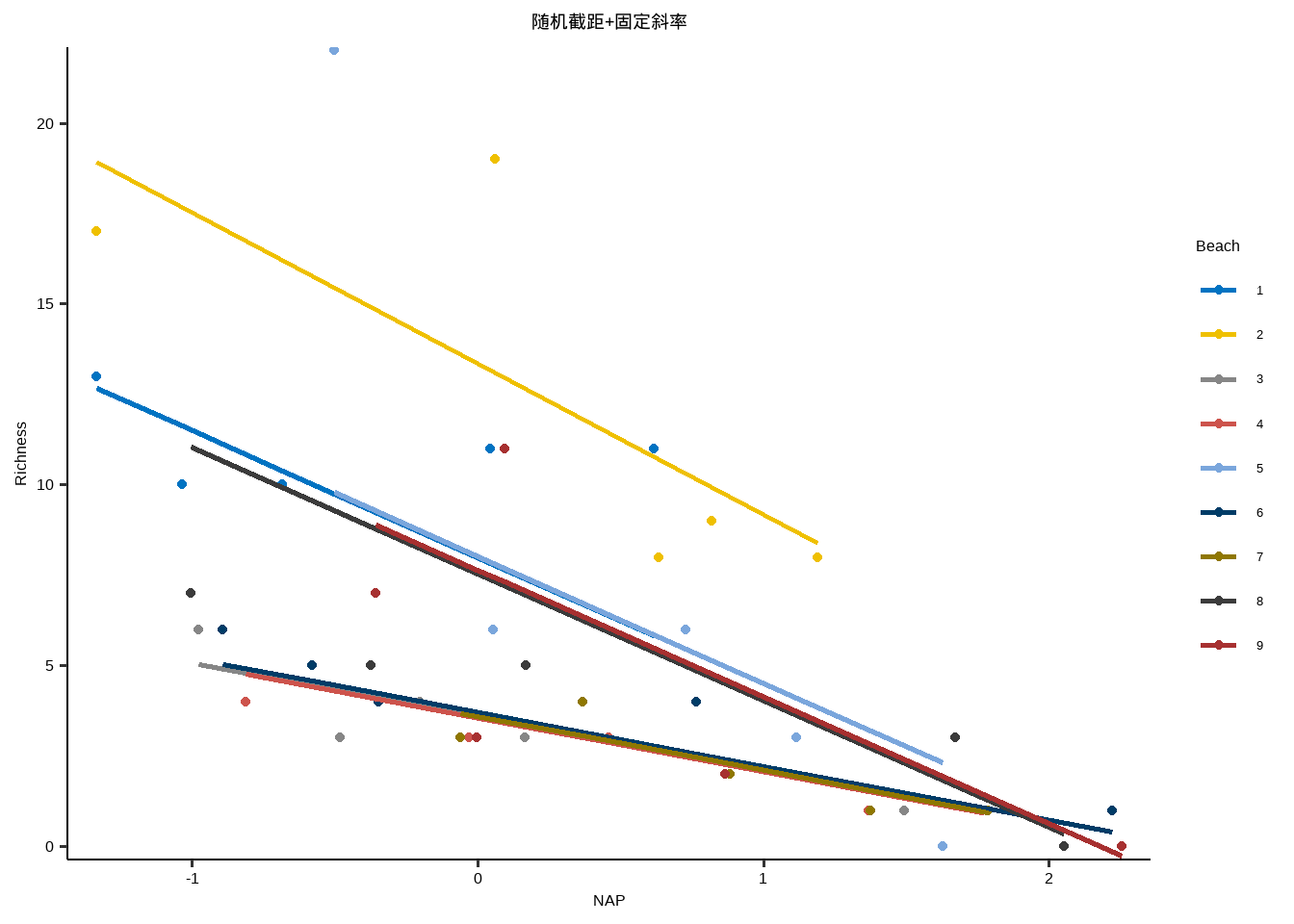
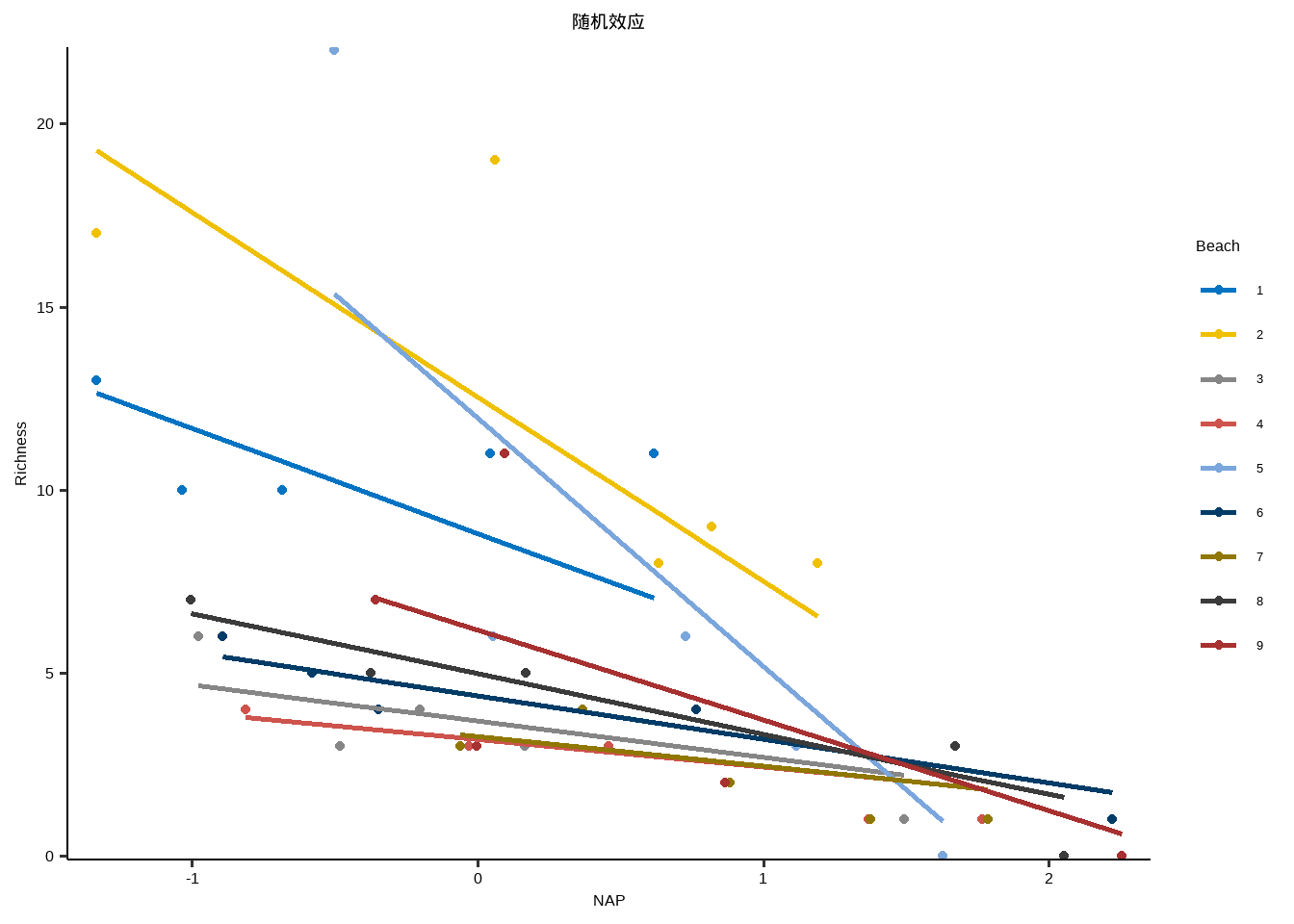
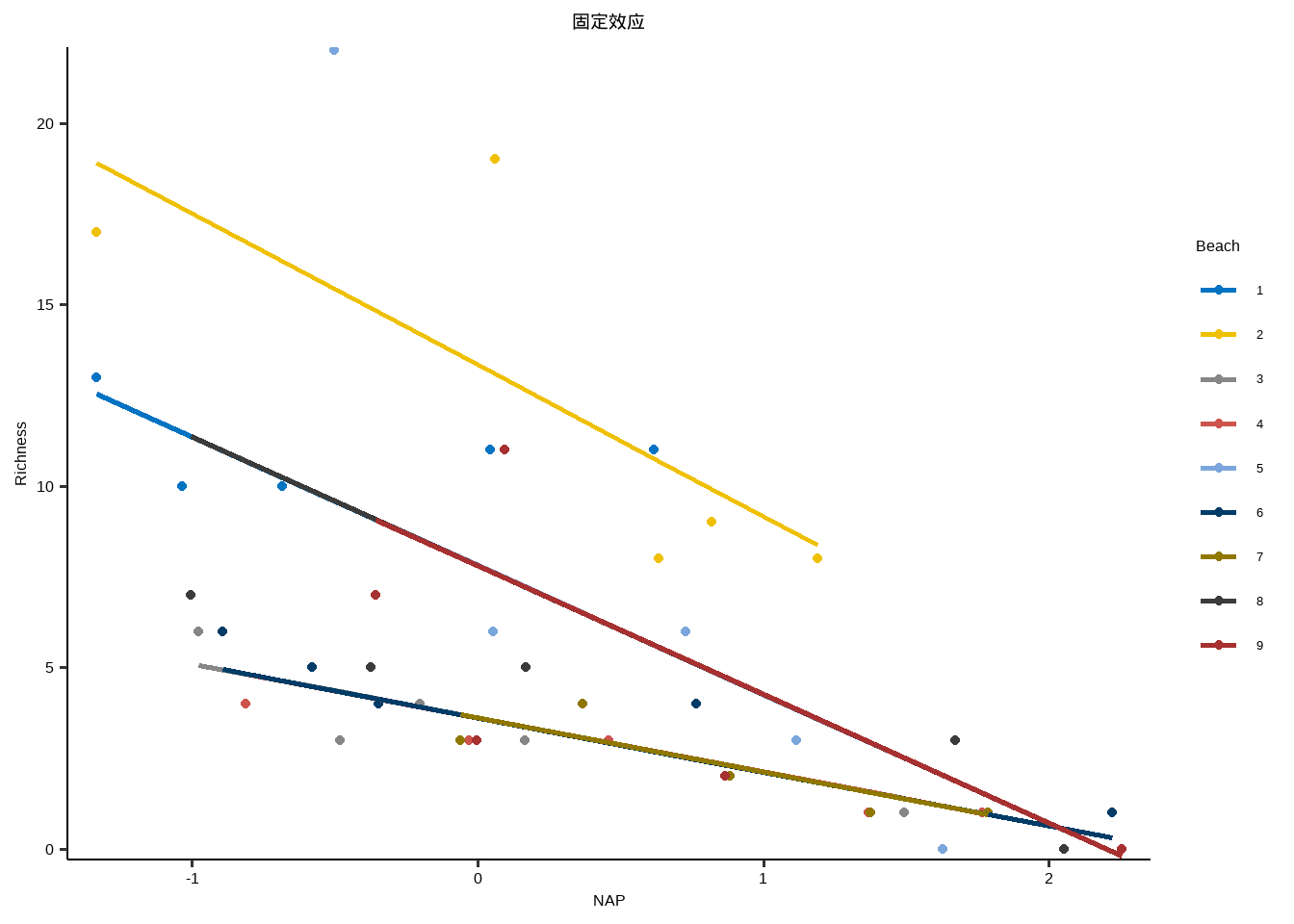
} library ( purrr ) library ( ggplot2 ) lme_plot <- map2 ( list ( lme1 ,lme2 ,lme3 , lme4 ,lm ) ,list ( "随机截距+随机斜率" ,"随机截距+固定斜率" ,"固定截距+随机斜率" ,"随机效应" ,"固定效应" ) ,plot_lme ) lme_plot #> [[1]] #>
#> [[2]]#>
#> [[3]]#>
#> [[4]]#>
#> [[5]]
Show the code anova ( lme1 ,lme2 ,lme3 ,lme4 ,lm ) #> Data: rikz #> Models: #> lme4: Richness ~ 1 + (1 + NAP | Beach) #> lm: Richness ~ 1 + NAP * Exposure #> lme2: Richness ~ 1 + NAP * Exposure + (1 + 1 | Beach) #> lme3: Richness ~ 1 + NAP * Exposure + (0 + NAP | Beach) #> lme1: Richness ~ 1 + NAP + Exposure + NAP:Exposure + (1 + NAP | Beach) #> npar AIC BIC logLik -2*log(L) Chisq Df Pr(>Chisq) #> lme4 5 254.24 263.27 -122.12 244.24 #> lm 7 234.26 246.91 -110.13 220.26 23.9789 2 6.209e-06 *** #> lme2 8 236.26 250.71 -110.13 220.26 0.0000 1 1.0000 #> lme3 8 236.26 250.71 -110.13 220.26 0.0000 0 #> lme1 10 238.20 256.27 -109.10 218.20 2.0619 2 0.3567 #> --- #> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 # p小于0.05,说明全模型与简化后模型存在差异,最终采用lme1,AIC
模型诊断
sleepstudy
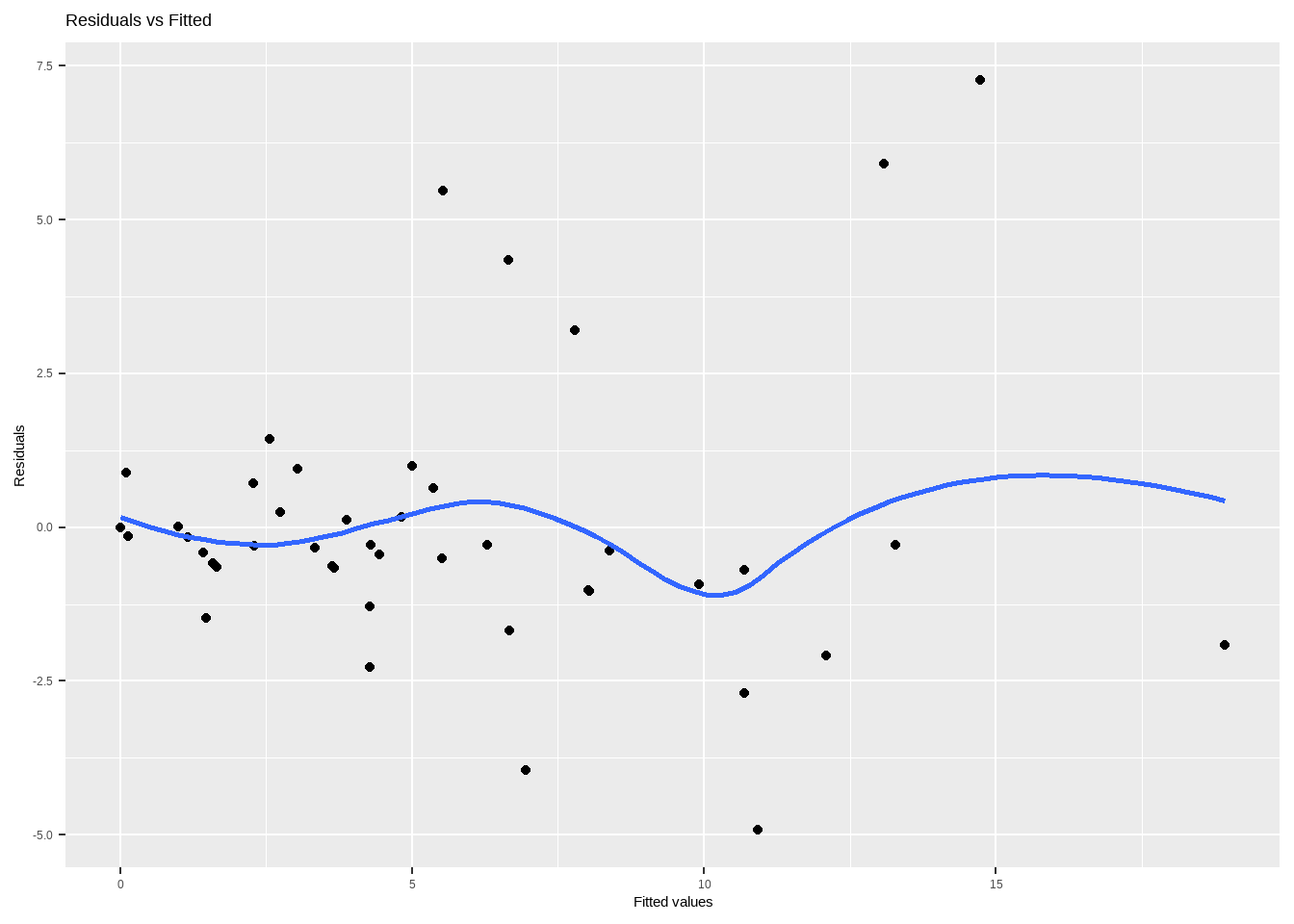
Show the code # 拟合线性混合模型 model <- lme1 # 1. 残差图 residuals <- resid ( model ) fitted <- fitted ( model ) ggplot ( data.frame ( fitted , residuals ) , aes ( fitted , residuals ) ) + geom_point ( ) +
geom_smooth ( method = "loess" , se = FALSE ) +
labs ( title = "Residuals vs Fitted" , x = "Fitted values" , y = "Residuals" ) Show the code
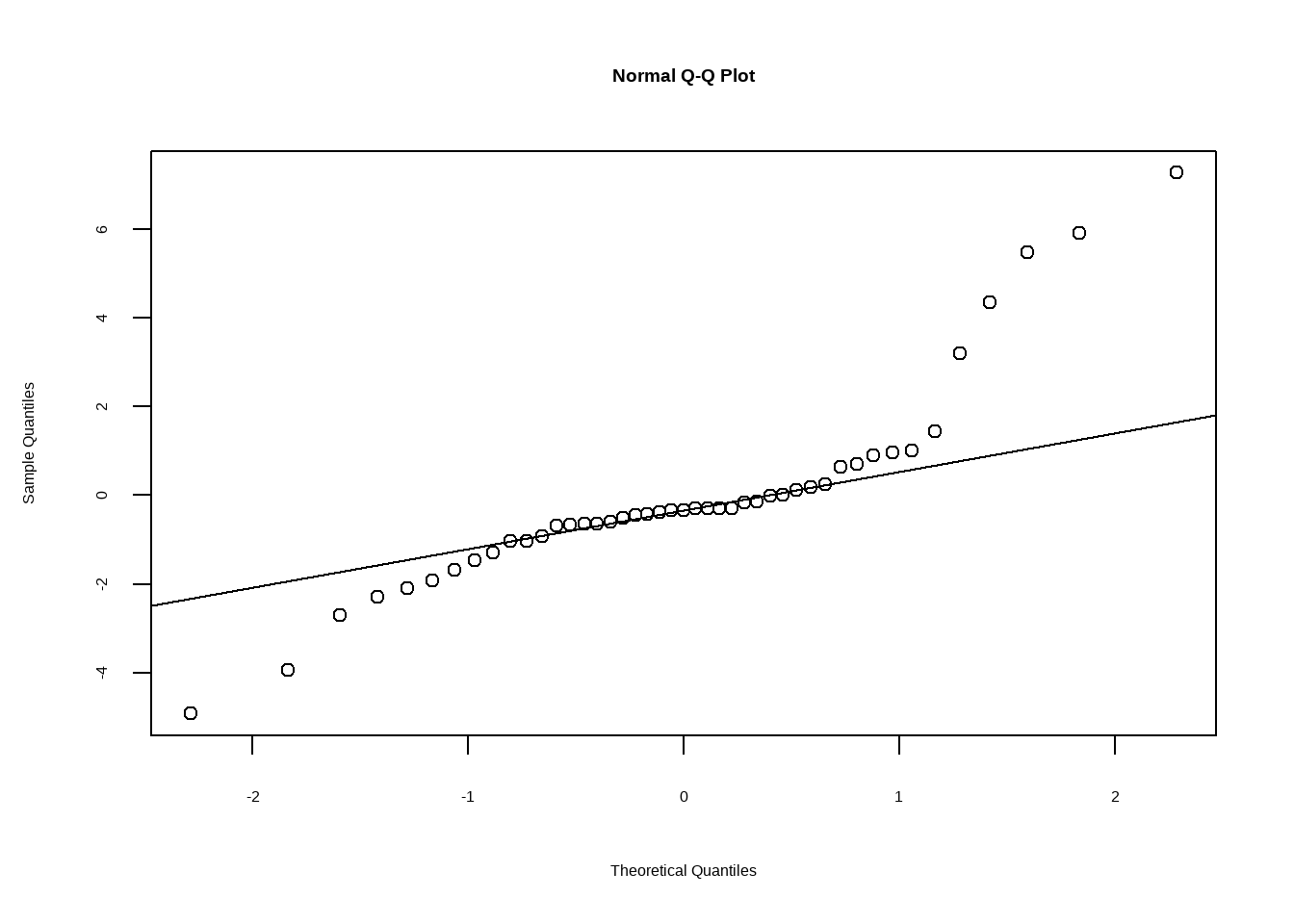
Show the code # 3. Cook's 距离 cooksd <- cooks.distance ( model ) plot ( cooksd , type = "h" , main = "Cook's Distance" ) Show the code # 4. 随机效应的分布 rand_dist <- ranef ( model ) qqnorm ( rand_dist $ Beach $ NAP ) qqline ( rand_dist $ Beach $ NAP )